1H5D
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (0-11% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, PEROXIDASE C1A, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-21 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1H5E
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (11-22% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, PEROXIDASE C1A, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-21 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1H5F
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (22-33% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-21 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1H5G
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (33-44% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-21 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1H5H
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (44-56% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-22 | | Release date: | 2002-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1H5I
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (56-67% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-22 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1H5J
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (67-78% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-22 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1H5K
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (78-89% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-22 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1H5L
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (89-100% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-22 | | Release date: | 2002-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1H5M
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (0-100% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-22 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1H5N
 
 | | DMSO REDUCTASE MODIFIED BY THE PRESENCE OF DMS AND AIR | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM(VI) ION, ... | | Authors: | Bailey, S, Adams, B, Smith, A.T, Richards, R.L, Lowe, D.J, Bray, R.C. | | Deposit date: | 2001-05-22 | | Release date: | 2002-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reactions of Dimethyl Sulphoxide Reductase in the Presence of Dimethylsulphide and the Structure of the Dimethylsulphide-Modified Enzyme
Biochemistry, 40, 2001
|
|
1H5O
 
 | | Solution structure of Crotamine, a neurotoxin from Crotalus durissus terrificus | | Descriptor: | MYOTOXIN | | Authors: | Nicastro, G, Franzoni, L, De Chiara, C, Mancin, C.A, Giglio, J.R, Spisni, A. | | Deposit date: | 2001-05-23 | | Release date: | 2003-05-09 | | Last modified: | 2013-07-24 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Crotamine, a Na+ Channel Affecting Toxin from Crotalus Durissus Terrificus Venom
Eur.J.Biochem., 270, 2003
|
|
1H5P
 
 | | Solution structure of the human Sp100b SAND domain by heteronuclear NMR. | | Descriptor: | NUCLEAR AUTOANTIGEN SP100-B | | Authors: | Bottomley, M.J, Liu, Z, Collard, M.W, Huggenvik, J.I, Gibson, T.J, Sattler, M. | | Deposit date: | 2001-05-24 | | Release date: | 2001-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation.
Nat. Struct. Biol., 8, 2001
|
|
1H5Q
 
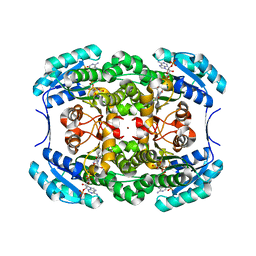 | | Mannitol dehydrogenase from Agaricus bisporus | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT MANNITOL DEHYDROGENASE, NICKEL (II) ION | | Authors: | Horer, S, Stoop, J, Mooibroek, H, Baumann, U, Sassoon, J. | | Deposit date: | 2001-05-24 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystallographic Structure of the Mannitol 2-Dehydrogenase Nadp+ Binary Complex from Agaricus Bisporus
J.Biol.Chem., 276, 2001
|
|
1H5R
 
 | | Thymidylyltransferase complexed with Thimidine and Glucose-1-phospate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION, ... | | Authors: | Rosano, C, Zuccotti, S, Bolognesi, M. | | Deposit date: | 2001-05-25 | | Release date: | 2001-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and Crystallographic Analyses Support a Sequential-Ordered Bi Bi Catalytic Mechanism for Escherichia Coli Glucose-1-Phosphate Thymidylyltransferase
J.Mol.Biol., 313, 2001
|
|
1H5S
 
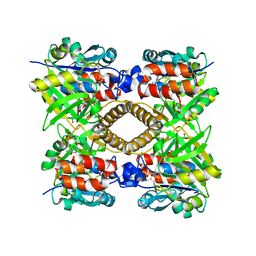 | | Thymidylyltransferase complexed with TMP | | Descriptor: | Glucose-1-phosphate thymidylyltransferase 1, THYMIDINE-5'-PHOSPHATE | | Authors: | Rosano, C, Zuccotti, S, Bolognesi, M. | | Deposit date: | 2001-05-25 | | Release date: | 2001-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and crystallographic analyses support a sequential-ordered bi bi catalytic mechanism for Escherichia coli glucose-1-phosphate thymidylyltransferase.
J. Mol. Biol., 313, 2001
|
|
1H5T
 
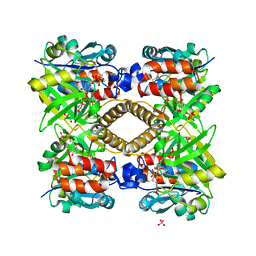 | | Thymidylyltransferase complexed with Thymidylyldiphosphate-glucose | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, Glucose-1-phosphate thymidylyltransferase 1, SULFATE ION, ... | | Authors: | Rosano, C, Zuccotti, S, Bolognesi, M. | | Deposit date: | 2001-05-25 | | Release date: | 2001-11-23 | | Last modified: | 2018-12-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and Crystallographic Analyses Support a Sequential-Ordered Bi Bi Catalytic Mechanism for Escherichia Coli Glucose-1-Phosphate Thymidylyltransferase
J.Mol.Biol., 313, 2001
|
|
1H5U
 
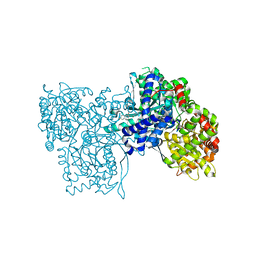 | | THE 1.76 A RESOLUTION CRYSTAL STRUCTURE OF GLYCOGEN PHOSPHORYLASE B COMPLEXED WITH GLUCOSE AND CP320626, A POTENTIAL ANTIDIABETIC DRUG | | Descriptor: | 5-CHLORO-1H-INDOLE-2-CARBOXYLIC ACID [1-(4-FLUOROBENZYL)-2-(4-HYDROXYPIPERIDIN-1YL)-2-OXOETHYL]AMIDE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Archontis, G. | | Deposit date: | 2001-05-25 | | Release date: | 2001-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The 1.76 A Resolution Crystal Structure of Glycogen Phosphorylase B Complexed with Glucose, and Cp320626, a Potential Antidiabetic Drug
Bioorg.Med.Chem., 10, 2002
|
|
1H5V
 
 | | Thiopentasaccharide complex of the endoglucanase Cel5A from Bacillus agaradharens at 1.1 A resolution in the tetragonal crystal form | | Descriptor: | CALCIUM ION, ENDOGLUCANASE 5A, SODIUM ION, ... | | Authors: | Varrot, A, Sulzenbacher, G, Schulein, M, Driguez, H, Davies, G.J. | | Deposit date: | 2001-05-28 | | Release date: | 2002-05-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structure of Endoglucanase Cel5A in Complex with Methyl 4,4II,4III,4Iv-Tetrathio-Alpha-Cellopentoside Highlights the Alternative Binding Modes Targeted by Substrate Mimics
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1H5W
 
 | | 2.1A Bacteriophage Phi-29 Connector | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, UPPER COLLAR PROTEIN | | Authors: | Guasch, A, Pous, J, Ibarra, B, Gomis-Ruth, F.X, Valpuesta, J.M, Sousa, N, Carrascosa, J.L, Coll, M. | | Deposit date: | 2001-05-28 | | Release date: | 2002-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detailed Architecture of a DNA Translocating Machine: The High-Resolution Structure of the Bacteriophage Phi29 Connector Particle.
J.Mol.Biol., 315, 2002
|
|
1H5X
 
 | |
1H5Y
 
 | | HisF protein from Pyrobaculum aerophilum | | Descriptor: | GLYCEROL, HISF, PHOSPHATE ION | | Authors: | Banfield, M.J, Lott, J.S, McCarthy, A.A, Baker, E.N. | | Deposit date: | 2001-05-31 | | Release date: | 2001-06-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Hisf, a Histidine Biosynthetic Protein from Pyrobaculum Aerophilum
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1H5Z
 
 | |
1H60
 
 | |
1H61
 
 | | Structure of Pentaerythritol Tetranitrate Reductase in complex with prednisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2001-06-04 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
