5FP8
 
 | |
2OD5
 
 | |
6Y0N
 
 | | Arginine hydroxylase VioC in complex with Arg, 2OG and Fe under anaerobic environment using FT-SSX methods | | Descriptor: | 2-OXOGLUTARIC ACID, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Rabe, P, Beale, J.H, Lang, P.A, Dirr, S.A, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Anaerobic fixed-target serial crystallography.
Iucrj, 7, 2020
|
|
5CSD
 
 | |
2OD4
 
 | |
8AY3
 
 | |
8AY8
 
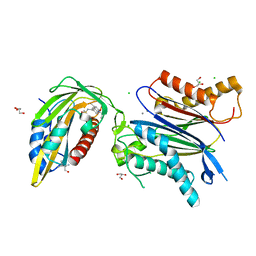 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1(V112L, T135L,F137I, T153I, V168A)-iSB9-HAB1 TERNARY COMPLEX | | Descriptor: | Abscisic acid receptor PYL1, CHLORIDE ION, GLYCEROL, ... | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
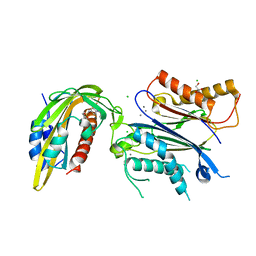
8AYA
 
 | |
8AY6
 
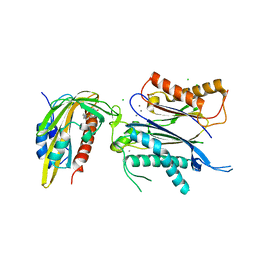 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1(V112L, T135L,F137I, T153I, V168A)-SB-HAB1 TERNARY COMPLEX | | Descriptor: | 1,4-dimethyl-2-oxidanylidene-~{N}-(phenylmethyl)quinoline-6-sulfonamide, Abscisic acid receptor PYL1, CHLORIDE ION, ... | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
8AY9
 
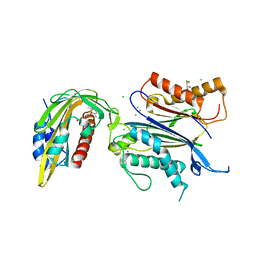 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1(V112L, T135L,F137I, T153I, V168A)-ABA-HAB1 TERNARY COMPLEX | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL1, CHLORIDE ION, ... | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
8AY7
 
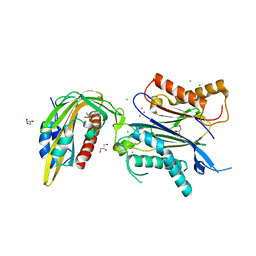 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1(V112L, T135L,F137I, T153I, V168A)-iSB7-HAB1 TERNARY COMPLEX | | Descriptor: | Abscisic acid receptor PYL1, CHLORIDE ION, GLYCEROL, ... | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.131 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
5DMY
 
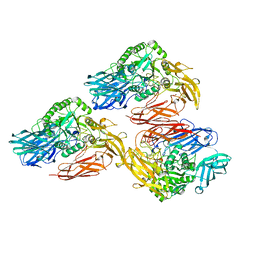 | | Beta-galactosidase - construct 33-930 | | Descriptor: | 1,2-ETHANEDIOL, Beta-galactosidase, MAGNESIUM ION, ... | | Authors: | Watson, K.A, Lazidou, A. | | Deposit date: | 2015-09-09 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational protein engineering toward the development of a beta-galactosidase with improved functional properties
To Be Published
|
|
5BZ5
 
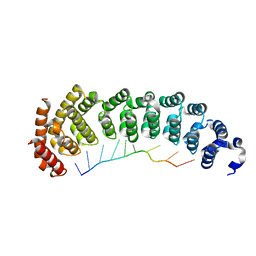 | |
5BZV
 
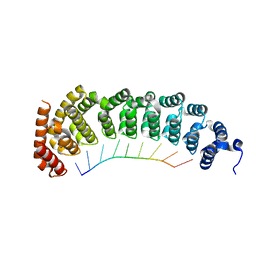 | |
5H8X
 
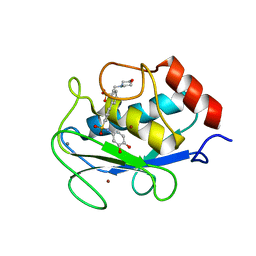 | | Crystal structure of the complex MMP-8/BF471 (catechol inhibitor) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Neutrophil collagenase, ... | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Tortorella, P. | | Deposit date: | 2015-12-24 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Catechol-based matrix metalloproteinase inhibitors with additional antioxidative activity.
J Enzyme Inhib Med Chem, 31, 2016
|
|
3WSA
 
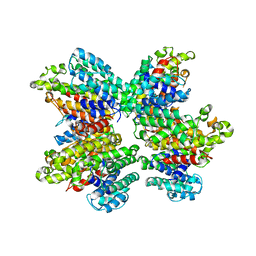 | | The Tuberculosis Drug SQ109 Inhibits Trypanosoma cruzi Cell Proliferation and acts Synergistically with Posaconazole | | Descriptor: | N-[(2Z)-3,7-dimethylocta-2,6-dien-1-yl]-N'-[(1R,3S,5R,7R)-tricyclo[3.3.1.1~3,7~]dec-2-yl]ethane-1,2-diamine, Squalene synthase | | Authors: | Shang, N, Li, Q, Huang, C.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-03-05 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | SQ109, a new drug lead for Chagas disease.
Antimicrob.Agents Chemother., 59, 2015
|
|
5BYM
 
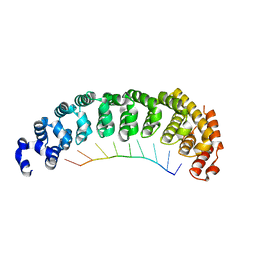 | |
3WCC
 
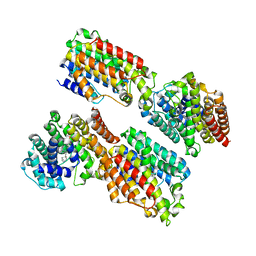 | | The complex structure of TcSQS with ligand, E5700 | | Descriptor: | (3R)-3-({2-benzyl-6-[(3R,4S)-3-hydroxy-4-methoxypyrrolidin-1-yl]pyridin-3-yl}ethynyl)-1-azabicyclo[2.2.2]octan-3-ol, Farnesyltransferase, putative | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
2PAJ
 
 | |
5AAN
 
 | | Crystal structure of Drosophila NCS-1 bound to penothiazine FD44 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, CG5907-PA, ... | | Authors: | Chaves-Sanjuan, A, Infantes, L, Sanchez-Barrena, M.J. | | Deposit date: | 2015-07-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interference of the complex between NCS-1 and Ric8a with phenothiazines regulates synaptic function and is an approach for fragile X syndrome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5HM0
 
 | | Crystal structure of the first bromodomain of human BRD4 bound to benzoisoxazoloazepine 3 | | Descriptor: | 6-(4-chlorophenyl)-1-methyl-4H-[1,2]oxazolo[5,4-d][2]benzazepine, Bromodomain-containing protein 4 | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-01-15 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Identification of a Benzoisoxazoloazepine Inhibitor (CPI-0610) of the Bromodomain and Extra-Terminal (BET) Family as a Candidate for Human Clinical Trials.
J.Med.Chem., 59, 2016
|
|
2PGC
 
 | |
3UW3
 
 | |
7X8A
 
 | | Cryo-EM structure of a bacterial protein complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7XC7
 
 | | Cryo-EM structure of a bacterial protein complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
