3AYX
 
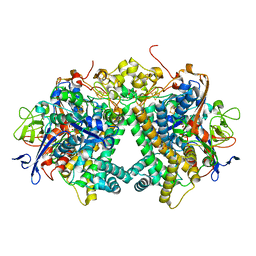 | | Membrane-bound respiratory [NiFe] hydrogenase from Hydrogenovibrio marinus in an H2-reduced condition | | Descriptor: | CARBON MONOXIDE, CYANIDE ION, FE (II) ION, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2011-05-20 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
3AYZ
 
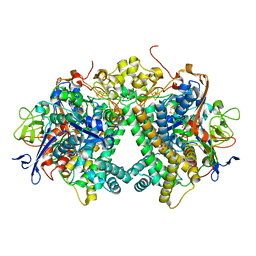 | | Membrane-bound respiratory [NiFe] hydrogenase from Hydrogenovibrio marinus in an air-oxidized condition | | Descriptor: | CARBON MONOXIDE, CYANIDE ION, FE (II) ION, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2011-05-20 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
1H2A
 
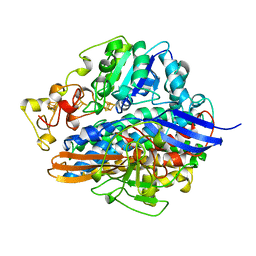 | | SINGLE CRYSTALS OF HYDROGENASE FROM DESULFOVIBRIO VULGARIS | | Descriptor: | FE3-S4 CLUSTER, HYDROGENASE, IRON/SULFUR CLUSTER, ... | | Authors: | Higuchi, Y, Yasuoka, N. | | Deposit date: | 1997-10-17 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual ligand structure in Ni-Fe active center and an additional Mg site in hydrogenase revealed by high resolution X-ray structure analysis.
Structure, 5, 1997
|
|
1H2R
 
 | |
3RGW
 
 | | Crystal structure at 1.5 A resolution of an H2-reduced, O2-tolerant hydrogenase from Ralstonia eutropha unmasks a novel iron-sulfur cluster | | Descriptor: | FE3-S4 CLUSTER, FE4-S3 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Scheerer, P, Fritsch, J, Frielingsdorf, S, Kroschinsky, S, Friedrich, B, Lenz, O, Spahn, C.M.T. | | Deposit date: | 2011-04-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre.
Nature, 479, 2011
|
|
3USC
 
 | | Crystal Structure of E. coli hydrogenase-1 in a ferricyanide-oxidized form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3CUR
 
 | | Structure of a double methionine mutant of NI-FE hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A. | | Deposit date: | 2008-04-17 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental approaches to kinetics of gas diffusion in hydrogenase
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3UQY
 
 | | H2-reduced structure of E. coli hydrogenase-1 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3USE
 
 | | Crystal Structure of E. coli hydrogenase-1 in its as-isolated form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5ODI
 
 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus cocrystallized with CoM-SH | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
5JRD
 
 | | E. coli Hydrogenase-1 variant P508A | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Armstrong, F.A, Evans, R.M, Brooke, E.J, Islam, S.T.A. | | Deposit date: | 2016-05-06 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Importance of the Active Site "Canopy" Residues in an O2-Tolerant [NiFe]-Hydrogenase.
Biochemistry, 56, 2017
|
|
5JSK
 
 | | The 3D structure of [NiFeSe] hydrogenase from Desulfovibrio vulgaris Hildenborough in the reduced state at 0.95 Angstrom resolution | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, GLYCEROL, ... | | Authors: | Marques, M.C, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The direct role of selenocysteine in [NiFeSe] hydrogenase maturation and catalysis.
Nat. Chem. Biol., 13, 2017
|
|
5JT1
 
 | | The 3D structure of Ni-reconstituted U489C variant of [NiFeSe] hydrogenase from Desulfovibrio vulgaris Hildenborough in the oxidized state at 1.35 Angstrom resolution | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, HYDROSULFURIC ACID, ... | | Authors: | Marques, M.C, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2016-05-09 | | Release date: | 2017-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The direct role of selenocysteine in [NiFeSe] hydrogenase maturation and catalysis.
Nat. Chem. Biol., 13, 2017
|
|
5JSH
 
 | | The 3D structure of recombinant [NiFeSe] hydrogenase from Desulfovibrio Vulgaris Hildenborough in the oxidized state at 1.30 Angstrom | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE (II) ION, ... | | Authors: | Marques, M.C, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The direct role of selenocysteine in [NiFeSe] hydrogenase maturation and catalysis.
Nat. Chem. Biol., 13, 2017
|
|
3ZE8
 
 | | 3D structure of the Ni-Fe-Se hydrogenase from D. vulgaris Hildenborough in the reduced state at 1.95 Angstroms | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE (II) ION, ... | | Authors: | Marques, M.C, Coelho, R, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Redox State-Dependent Changes in the Crystal Structure of [Nifese] Hydrogenase from Desulfovibrio Vulgaris Hildenborough
Int.J.Hydrogen Energy, 38, 2013
|
|
5JSY
 
 | | The 3D structure of the Ni-reconstituted U489C variant of [NiFeSe] hydrogenase from Desulfovibrio vulgaris Hildenborough at 1.04 Angstrom resolution | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE (II) ION, ... | | Authors: | Marques, M.C, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2016-05-09 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | The direct role of selenocysteine in [NiFeSe] hydrogenase maturation and catalysis.
Nat. Chem. Biol., 13, 2017
|
|
3ZE6
 
 | | 3D structure of the Ni-Fe-Se hydrogenase from D. vulgaris Hildenborough in the as-isolated oxidized state at 1.50 Angstroms | | Descriptor: | 3-[DODECYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, BIS-(MU-2-OXO),[(MU-3--SULFIDO)-BIS(MU-2--SULFIDO)-TRIS(CYS-S)-TRI-IRON] (AQUA)(GLU-O)IRON(II), CARBONMONOXIDE-(DICYANO) IRON, ... | | Authors: | Marques, M.C, Coelho, R, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox State-Dependent Changes in the Crystal Structure of [Nifese] Hydrogenase from Desulfovibrio Vulgaris Hildenborough
Int.J.Hydrogen Energy, 2013
|
|
5ODQ
 
 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus soaked with bromoethanesulfonate. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-bromanylethanesulfonic acid, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-06 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
5ODC
 
 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus at 2.3 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
5JSU
 
 | | The 3D structure of the U489C variant of [NiFeSe] hydrogenase from Desulfovibrio vulgaris Hildenborough in the oxidized state at 1.40 Angstrom resolution | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, HYDROSULFURIC ACID, ... | | Authors: | Marques, M.C, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2016-05-09 | | Release date: | 2017-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The direct role of selenocysteine in [NiFeSe] hydrogenase maturation and catalysis.
Nat. Chem. Biol., 13, 2017
|
|
3ZFS
 
 | | Cryo-EM structure of the F420-reducing NiFe-hydrogenase from a methanogenic archaeon with bound substrate | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, COENZYME F420, F420-REDUCING HYDROGENASE, ... | | Authors: | Mills, D.J, Vitt, S, Strauss, M, Shima, S, Vonck, J. | | Deposit date: | 2012-12-12 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | De Novo Modeling of the F420-Reducing [Nife]-Hydrogenase from a Methanogenic Archaeon by Cryo-Electron Microscopy
Elife, 2, 2013
|
|
6I0D
 
 | | Respiratory complex I from Thermus thermophilus with bound Decyl-Ubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2018-10-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6HUM
 
 | | Structure of the photosynthetic complex I from Thermosynechococcus elongatus | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, IRON/SULFUR CLUSTER, ... | | Authors: | Schuller, J.M, Schuller, S.K, Kurisu, G, Engel, B.D, Nowaczyk, M.M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer.
Science, 363, 2019
|
|
5O31
 
 | | Mitochondrial complex I in the deactive state | | Descriptor: | Acyl carrier protein, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Blaza, J.N, Vinothkumar, K.R, Hirst, J. | | Deposit date: | 2017-05-23 | | Release date: | 2018-01-17 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Structure of the Deactive State of Mammalian Respiratory Complex I.
Structure, 26, 2018
|
|
5ODH
 
 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus soaked with heterodisulfide for 3.5 minutes | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
