6O0L
 
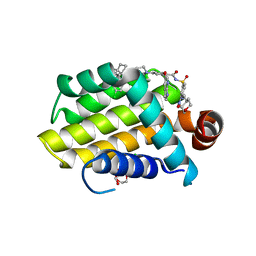 | | crystal structure of BCL-2 G101V mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, CHLORIDE ION, ... | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O0P
 
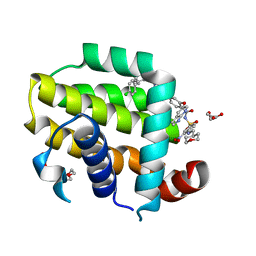 | | crystal structure of BCL-2 G101A mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, DI(HYDROXYETHYL)ETHER | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
2X0R
 
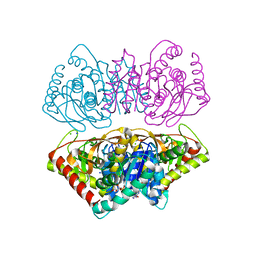 | | R207S, R292S Mutant of Malate Dehydrogenase from the Halophilic Archeon Haloarcula marismortui (HoloForm) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Irimia, A, Ebel, C, Vellieux, F.M.D, Richard, S.B, Cosenza, L.W, Zaccai, G, Madern, D. | | Deposit date: | 2009-12-17 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.915 Å) | | Cite: | The Oligomeric States of Haloarcula Marismortui Malate Dehydrogenase are Modulated by Solvent Components as Shown by Crystallographic and Biochemical Studies
J.Mol.Biol., 326, 2003
|
|
3L4Y
 
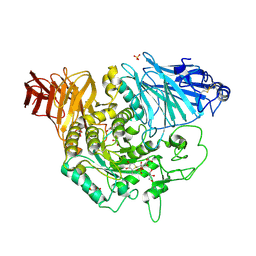 | | Crystal complex of N-terminal Human Maltase-Glucoamylase with NR4-8II | | Descriptor: | (1S,2R,3S,4R)-1-{(1S)-2-[(2R,3S,4S)-3,4-dihydroxy-2-(hydroxymethyl)tetrahydrothiophenium-1-yl]-1-hydroxyethyl}-2,3,4,5-tetrahydroxypentyl sulfate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata.
Biochemistry, 49, 2010
|
|
4N7L
 
 | | Zinc Substituted Reaction Center M(L214H) Variant of Rhodobacter sphaeroides | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, FE (III) ION, ... | | Authors: | Hardjasa, A, Murphy, M.E.P. | | Deposit date: | 2013-10-15 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and kinetic properties of Rhodobacter sphaeroides photosynthetic reaction centers containing exclusively Zn-coordinated bacteriochlorophyll as bacteriochlorin cofactors.
Biochim.Biophys.Acta, 1837, 2014
|
|
3L4X
 
 | | Crystal complex of N-terminal Human Maltase-Glucoamylase with NR4-8 | | Descriptor: | (1S,2R,3S,4S)-1-{(1S)-2-[(2R,3S,4S)-3,4-dihydroxy-2-(hydroxymethyl)tetrahydrothiophenium-1-yl]-1-hydroxyethyl}-2,3,4,5-tetrahydroxypentyl sulfate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata.
Biochemistry, 49, 2010
|
|
4N7K
 
 | | Zinc Substituted Reaction Center of the Rhodobacter sphaeroides | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, FE (III) ION, ... | | Authors: | Hardjasa, A, Murphy, M.E.P. | | Deposit date: | 2013-10-15 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and kinetic properties of Rhodobacter sphaeroides photosynthetic reaction centers containing exclusively Zn-coordinated bacteriochlorophyll as bacteriochlorin cofactors.
Biochim.Biophys.Acta, 1837, 2013
|
|
1T8U
 
 | | Crystal Structure of human 3-O-Sulfotransferase-3 with bound PAP and tetrasaccharide substrate | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Moon, A.F, Edavettal, S.C, Krahn, J.M, Munoz, E.M, Negishi, M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of the sulfotransferase (3-o-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1
J.Biol.Chem., 279, 2004
|
|
2QE8
 
 | |
6TTS
 
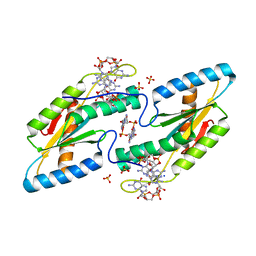 | | Crystal structure of the GGDEF domain of DgcB from Caulobacter crescentus in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GGDEF diguanylate cyclase DgcB, SULFATE ION | | Authors: | Holzschuh, F, Schirmer, T, Teixeira, R. | | Deposit date: | 2019-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the GGDEF domain of DgcB from Caulobacter crescentus in complex with c-di-GMP
To Be Published
|
|
8GTM
 
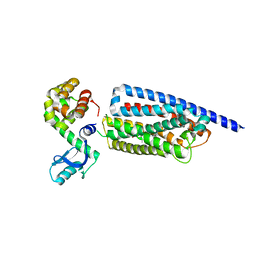 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C203 by XFEL | | Descriptor: | 7-(4-bromanyl-2,6-dimethoxy-phenyl)-4,8-dimethyl-~{N},~{N}-bis[4,4,4-tris(fluoranyl)butyl]-1$l^{4},3,5,9-tetrazabicyclo[4.3.0]nona-1(6),2,4,8-tetraen-2-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
6U0S
 
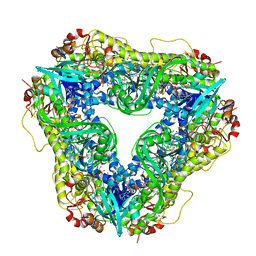 | | Crystal structure of the flavin-dependent monooxygenase PieE in complex with FAD and substrate | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, 2-[(2E,5E,7E,9R,10R,11E)-10-hydroxy-3,7,9,11-tetramethyltrideca-2,5,7,11-tetraen-1-yl]-6-methoxy-3-methylpyridin-4-ol, CHLORIDE ION, ... | | Authors: | Shi, R, Manenda, M. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
6A04
 
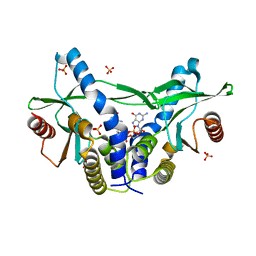 | | Structure of pSTING complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
7XQW
 
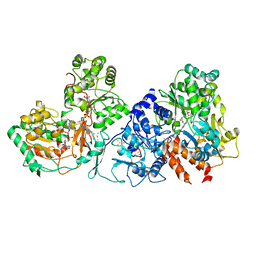 | | Formate dehydrogenase (FDH) from Methylobacterium extorquens AM1 (MeFDH1) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Park, J, Heo, Y.Y, Roh, S.H, Lee, H.H. | | Deposit date: | 2022-05-09 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Enzymatic conversion of CO2 in real flue gas to molar-scale formate
To Be Published
|
|
7Y61
 
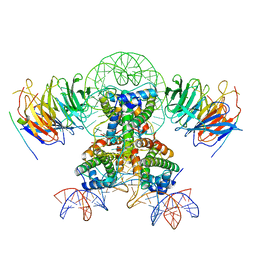 | | Cryo-EM structure of the two CAF1LCs bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5W
 
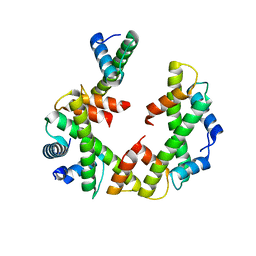 | | Cryo-EM structure of the left-handed Di-tetrasome | | Descriptor: | Histone H3.1, Histone H4, Widom 601 DNA (147-MER) | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y60
 
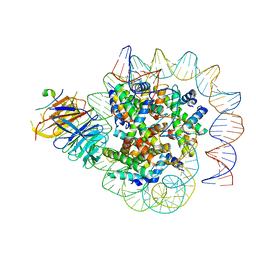 | | Cryo-EM structure of human CAF1LC bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2022-06-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7RK3
 
 | |
8T93
 
 | | Crystal structure of Terrestrivirus Inositol pyrophosphatase kinase in complex with ADP and scyllo-L-(1,2,3,4)-IP4 | | Descriptor: | (1R,2S,3S,4R,5S,6S)-5,6-dihydroxycyclohexane-1,2,3,4-tetrayl tetrakis[dihydrogen (phosphate)], ADENOSINE-5'-DIPHOSPHATE, Kinase, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
5XBT
 
 | | The structure of BrlR bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Wang, F, Qing, H, Gu, L. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin.
Nat Commun, 9, 2018
|
|
8T92
 
 | | Crystal structure of Terrestrivirus Inositol pyrophosphatase kinase in complex with ADP and scyllo-D-(1,2,3,4)-IP4 | | Descriptor: | (1S,2R,3R,4S,5R,6R)-5,6-dihydroxycyclohexane-1,2,3,4-tetrayl tetrakis[dihydrogen (phosphate)], ADENOSINE-5'-DIPHOSPHATE, Kinase, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
7R49
 
 | | Crystal structure of the L. plantarum acyl carrier protein synthase (AcpS)in complex with D-alanyl carrier protein (DltC1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4'-PHOSPHOPANTETHEINE, D-alanyl carrier protein 1, ... | | Authors: | Nikolopoulos, N, Ravaud, S, Simorre, J.P, Grangeasse, C. | | Deposit date: | 2022-02-08 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | DltC acts as an interaction hub for AcpS, DltA and DltB in the teichoic acid D-alanylation pathway of Lactiplantibacillus plantarum.
Sci Rep, 12, 2022
|
|
5XJW
 
 | | Crystal Structure of the [Co2+-(Chromomycin A3)2]-CCG repeats Complex | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-4-O-acetyl-2,6-dideoxy-beta-D-galactopyranose, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-beta-D-Olivopyranose-(1-3)-beta-D-Olivopyranose, ... | | Authors: | Tseng, W.H, Wu, P.C, Satange, R.B, Hou, M.H. | | Deposit date: | 2017-05-04 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal Structure of the [Co2+-(Chromomycin A3)2]-CCG repeats Complex
To be published
|
|
1A6H
 
 | |
6IBO
 
 | | Catalytic deficiency of O-GlcNAc transferase leads to X-linked intellectual disability | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, ALA-VAL-SER-ARG-ALA, PHOSPHATE ION, ... | | Authors: | Gundogdu, M, van Aalten, D.M.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Catalytic deficiency of O-GlcNAc transferase leads to X-linked intellectual disability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
