7MUB
 
 | | KcsA Open gate E71V mutant in Potassium | | Descriptor: | Fab heavy chain, Fab light chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Li, J, Weingarth, M, Roux, B. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A distinct mechanism of C-type inactivation in the Kv-like KcsA mutant E71V.
Nat Commun, 13, 2022
|
|
7MK6
 
 | | KcsA open gate E71V mutant with sodium | | Descriptor: | Fab heavy chain, Fab light chain, pH-gated potassium channel KcsA | | Authors: | Rohaim, A, Li, J, Weingarth, M, Roux, B. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A distinct mechanism of C-type inactivation in the Kv-like KcsA mutant E71V.
Nat Commun, 13, 2022
|
|
7MJT
 
 | | KcsA open gate E71V mutant with Barium | | Descriptor: | BARIUM ION, Fab heavy chain, Fab light chain, ... | | Authors: | Rohaim, A, Li, J, Weingarth, M, Roux, B. | | Deposit date: | 2021-04-20 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A distinct mechanism of C-type inactivation in the Kv-like KcsA mutant E71V.
Nat Commun, 13, 2022
|
|
6QZ4
 
 | | Structure of MHETase from Ideonella sakaiensis | | Descriptor: | CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase, SULFATE ION | | Authors: | Allen, M.D, Johnson, C.W, Knott, B.C, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2019-03-11 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and engineering of a two-enzyme system for plastics depolymerization.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6QOX
 
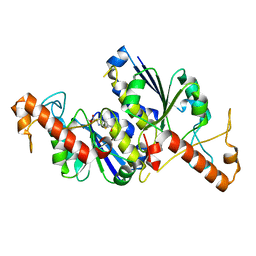 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 27 (Methyl 2-(hydroxymethyl)-6H-thieno[2,3-b]pyrrole-5-carboxylate) | | Descriptor: | methyl 2-(hydroxymethyl)-6~{H}-thieno[2,3-b]pyrrole-5-carboxylate, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
5LA6
 
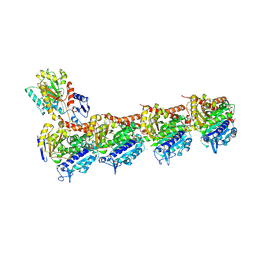 | | Tubulin-pironetin complex | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Prota, A.E, Setter, J, Waight, A.B, Bargsten, K, Murga, J, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pironetin Binds Covalently to alpha Cys316 and Perturbs a Major Loop and Helix of alpha-Tubulin to Inhibit Microtubule Formation.
J.Mol.Biol., 428, 2016
|
|
4K5Z
 
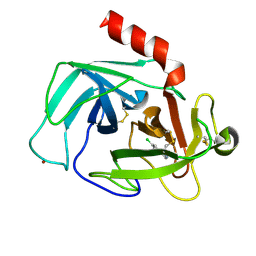 | | Crystal Structure of Human Chymase in Complex with Fragment Inhibitor 6-chloro-2,3-dihydro-1H-isoindol-1-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-2,3-dihydro-1H-isoindol-1-one, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
8B8O
 
 | |
1RVT
 
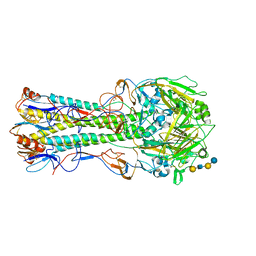 | | 1930 H1 Hemagglutinin in complex with LSTC | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Skehel, J.J, Gamblin, S.J, Haire, L.F, Russell, R.J, Stevens, D.J, Xiao, B, Ha, Y, Vasisht, N, Steinhauer, D.A, Daniels, R.S. | | Deposit date: | 2003-12-15 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure and receptor binding properties of the 1918 influenza hemagglutinin.
Science, 303, 2004
|
|
5WA9
 
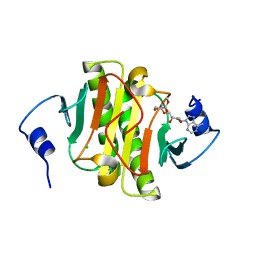 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside D-Ala phosphoramidate substrate complex | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[(2~{R})-1-methoxy-1-oxidanylidene-propan-2-yl]phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2017-06-26 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
6QWO
 
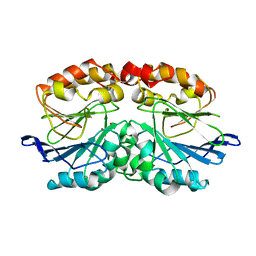 | | Zinc-reconstituted ODP from T. maritima | | Descriptor: | Oxygen-binding diiron protein, ZINC ION | | Authors: | Muok, A.R, Crane, B.R. | | Deposit date: | 2019-03-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A di-iron protein recruited as an Fe[II] and oxygen sensor for bacterial chemotaxis functions by stabilizing an iron-peroxy species.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8BD0
 
 | |
6WE4
 
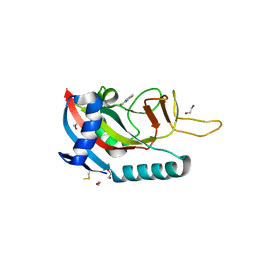 | | Human PARP14 (ARTD8), catalytic fragment in complex with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one, 8-methyl-2-{[(pyridin-4-yl)sulfanyl]methyl}quinazolin-4(3H)-one, ... | | Authors: | Swinger, K.S, Schenkel, L.B, Kuntz, K.W. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants.
Cell Chem Biol, 28, 2021
|
|
1RF1
 
 | | Crystal Structure of Fragment D of gammaE132A Fibrinogen with the Peptide Ligand Gly-His-Arg-Pro-amide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibrinogen alpha/alpha-E chain, ... | | Authors: | Kostelansky, M.S, Gorkun, O.V, Lord, S.T. | | Deposit date: | 2003-11-07 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Calcium-Binding Site beta2, Adjacent to the "b" Polymerization Site, Modulates Lateral Aggregation of Protofibrils during Fibrin Polymerization.
Biochemistry, 43, 2004
|
|
6WPM
 
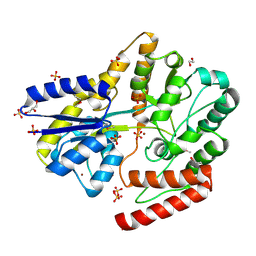 | | Crystal structure of a putative oligosaccharide periplasmic-binding protein from Synechococcus sp. MITs9220 in complex with zinc | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Ford, B.A, Michie, K.A, Paulsen, I.T, Mabbutt, B.C, Shah, B.S. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Novel functional insights into a modified sugar-binding protein from Synechococcus MITS9220.
Sci Rep, 12, 2022
|
|
6X58
 
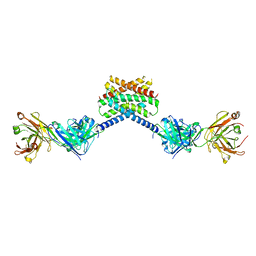 | | MPER-Fluc-Ec2 bound to 10E8v4 antibody | | Descriptor: | 10E8v4 Fab Heavy Chain, 10E8v4 Fab Light Chain, gp41 MPER peptide,Putative fluoride ion transporter CrcB | | Authors: | McIlwain, B.C, Stockbridge, R.B. | | Deposit date: | 2020-05-25 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | N-terminal Transmembrane-Helix Epitope Tag for X-ray Crystallography and Electron Microscopy of Small Membrane Proteins.
J.Mol.Biol., 433, 2021
|
|
1RSR
 
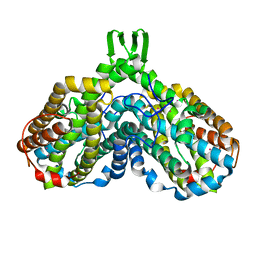 | | azide complex of the diferrous F208A mutant R2 subunit of ribonucleotide reductase | | Descriptor: | AZIDE ION, FE (II) ION, MERCURY (II) ION, ... | | Authors: | Andersson, M.E, Hogbom, M, Rinaldo-Matthis, A, Andersson, K.K, Sjoberg, B.M, Nordlund, P. | | Deposit date: | 2003-12-10 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of an Azide Complex of the Diferrous R2 Subunit of Ribonucleotide Reductase Displays a Novel Carboxylate Shift with Important Mechanistic Implications for Diiron-Catalyzed Oxygen Activation
J.Am.Chem.Soc., 121, 1999
|
|
5LZH
 
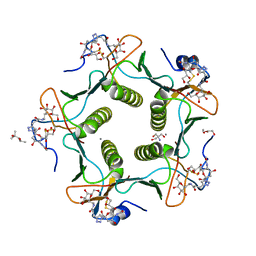 | | Cholera toxin classical B-pentamer in complex with inhibitor PC262 | | Descriptor: | (2~{R},4~{S},5~{R},6~{R})-5-acetamido-2-[4-[3-[2-[(2~{S},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]ethylamino]-3-oxidanylidene-propyl]-1,2,3-triazol-1-yl]-4-oxidanyl-6-[(1~{R},2~{R})-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Heggelund, J.E, Martinsen, T, Krengel, U. | | Deposit date: | 2016-09-29 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Towards new cholera prophylactics and treatment: Crystal structures of bacterial enterotoxins in complex with GM1 mimics.
Sci Rep, 7, 2017
|
|
6DNU
 
 | |
8T5S
 
 | | Cryo-EM structure of DRH-1 helicase and C-terminal domain bound to dsRNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-related helicase, MAGNESIUM ION, ... | | Authors: | Consalvo, C.D, Donelick, H.M, Shen, P.S, Bass, B.L. | | Deposit date: | 2023-06-14 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Caenorhabditis elegans Dicer acts with the RIG-I-like helicase DRH-1 and RDE-4 to cleave dsRNA.
Elife, 13, 2024
|
|
6WK5
 
 | |
6DA6
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes, apo form at 2.6 A resolution (P212121) | | Descriptor: | GLYCEROL, MAGNESIUM ION, UNKNOWN LIGAND, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
4JYJ
 
 | | Crystal Structure of putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans DSM 12444 | | Descriptor: | Enoyl-CoA hydratase/isomerase, UNKNOWN LIGAND | | Authors: | Cooper, D.R, Porebski, P.J, Domagalski, M.J, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans DSM 12444
To be Published
|
|
6QJU
 
 | |
1QKG
 
 | | DNA DECAMER DUPLEX CONTAINING T-T DEWAR PHOTOPRODUCT | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*(HYD)TP*+TP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*TP*GP*CP*G)-3') | | Authors: | Lee, J.-H, Bae, S.-H, Choi, Y.-J, Choi, B.-S. | | Deposit date: | 1999-07-20 | | Release date: | 2000-05-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Dewar Photoproduct of Thymidylyl(3'-->5')-Thymidine (Dewar Product) Exhibits Mutagenic Behavior in Accordance with the "A Rule".
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
