3DPA
 
 | |
1L41
 
 | |
3TIM
 
 | |
3TMS
 
 | |
1L48
 
 | |
1L51
 
 | |
1L60
 
 | |
1L67
 
 | |
4BP2
 
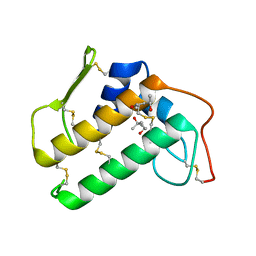 | | CRYSTALLOGRAPHIC REFINEMENT OF BOVINE PRO-PHOSPHOLIPASE A2 AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Finzel, B.C, Weber, P.C, Ohlendorf, D.H, Salemme, F.R. | | Deposit date: | 1990-09-07 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic refinement of bovine pro-phospholipase A2 at 1.6 A resolution.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
1L74
 
 | |
1L37
 
 | |
1L56
 
 | |
1SH1
 
 | |
5RUB
 
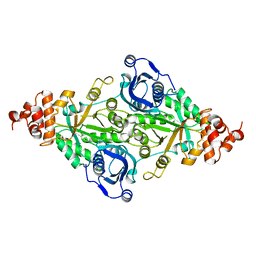 | | CRYSTALLOGRAPHIC REFINEMENT AND STRUCTURE OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE FROM RHODOSPIRILLUM RUBRUM AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | RUBISCO (RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE) | | Authors: | Schneider, G, Lindqvist, Y, Lundqvist, T. | | Deposit date: | 1990-05-29 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic refinement and structure of ribulose-1,5-bisphosphate carboxylase from Rhodospirillum rubrum at 1.7 A resolution.
J.Mol.Biol., 211, 1990
|
|
1L49
 
 | |
1L54
 
 | |
1L68
 
 | |
1LAP
 
 | | MOLECULAR STRUCTURE OF LEUCINE AMINOPEPTIDASE AT 2.7-ANGSTROMS RESOLUTION | | Descriptor: | Cytosol aminopeptidase, ZINC ION | | Authors: | Burley, S.K, David, P.R, Taylor, A, Lipscomb, W.N. | | Deposit date: | 1990-08-01 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular structure of leucine aminopeptidase at 2.7-A resolution.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
1L43
 
 | |
1L61
 
 | |
1L53
 
 | |
1L66
 
 | |
2GCT
 
 | |
2TRX
 
 | | CRYSTAL STRUCTURE OF THIOREDOXIN FROM ESCHERICHIA COLI AT 1.68 ANGSTROMS RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, THIOREDOXIN | | Authors: | Katti, S.K, Lemaster, D.M, Eklund, H. | | Deposit date: | 1990-03-19 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of thioredoxin from Escherichia coli at 1.68 A resolution.
J.Mol.Biol., 212, 1990
|
|
1FXI
 
 | |
