1EDK
 
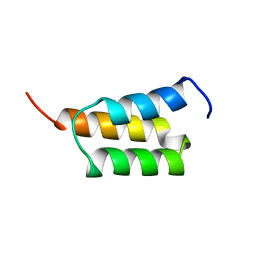 | |
1EDL
 
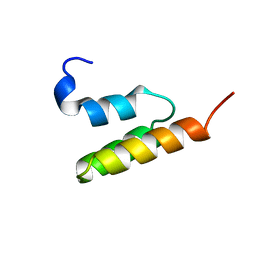 | |
1EDM
 
 | | EPIDERMAL GROWTH FACTOR-LIKE DOMAIN FROM HUMAN FACTOR IX | | Descriptor: | CALCIUM ION, FACTOR IX | | Authors: | Rao, Z, Handford, P, Mayhew, M, Knott, V, Brownlee, G.G, Stuart, D. | | Deposit date: | 1996-03-21 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of a Ca(2+)-binding epidermal growth factor-like domain: its role in protein-protein interactions.
Cell(Cambridge,Mass.), 82, 1995
|
|
1EDN
 
 | | HUMAN ENDOTHELIN-1 | | Descriptor: | ENDOTHELIN-1 | | Authors: | Wallace, B.A, Janes, R.W. | | Deposit date: | 1994-09-19 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The crystal structure of human endothelin.
Nat.Struct.Biol., 1, 1994
|
|
1EDO
 
 | | THE X-RAY STRUCTURE OF BETA-KETO ACYL CARRIER PROTEIN REDUCTASE FROM BRASSICA NAPUS COMPLEXED WITH NADP+ | | Descriptor: | BETA-KETO ACYL CARRIER PROTEIN REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Fisher, M, Kroon, J.T, Martindale, W, Stuitje, A.R, Slabas, A.R, Rafferty, J.B. | | Deposit date: | 2000-01-28 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray structure of Brassica napus beta-keto acyl carrier protein reductase and its implications for substrate binding and catalysis.
Structure Fold.Des., 8, 2000
|
|
1EDP
 
 | |
1EDQ
 
 | |
1EDR
 
 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO6AATTCGCG) AT 1.6 ANGSTROM | | Descriptor: | 5'-D(*CP*GP*CP*GP*(A47)AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION, SPERMINE | | Authors: | Chatake, T, Hikima, T, Ono, A, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2000-01-28 | | Release date: | 2000-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic studies on damaged DNAs. II. N(6)-methoxyadenine can present two alternate faces for Watson-Crick base-pairing, leading to pyrimidine transition mutagenesis.
J.Mol.Biol., 294, 1999
|
|
1EDS
 
 | | SOLUTION STRUCTURE OF INTRADISKAL LOOP 1 OF BOVINE RHODOPSIN (RHODOPSIN RESIDUES 92-123) | | Descriptor: | RHODOPSIN | | Authors: | Yeagle, P.L, Salloum, A, Chopra, A, Bhawsar, N, Ali, L. | | Deposit date: | 2000-01-28 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the intradiskal loops and amino terminus of the G-protein receptor, rhodopsin.
J.Pept.Res., 55, 2000
|
|
1EDT
 
 | |
1EDU
 
 | | CRYSTAL STRUCTURE OF THE ENTH DOMAIN OF RAT EPSIN 1 | | Descriptor: | 1,2-ETHANEDIOL, EH domain binding protein EPSIN | | Authors: | Hyman, J.H, Chen, H, Decamilli, P, Brunger, A.T. | | Deposit date: | 2000-01-28 | | Release date: | 2000-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Epsin 1 undergoes nucleocytosolic shuttling and its eps15 interactor NH(2)-terminal homology (ENTH) domain, structurally similar to Armadillo and HEAT repeats, interacts with the transcription factor promyelocytic leukemia Zn(2)+ finger protein (PLZF).
J.Cell Biol., 149, 2000
|
|
1EDV
 
 | | SOLUTION STRUCTURE OF 2ND INTRADISKAL LOOP OF BOVINE RHODOPSIN (RESIDUES 172-205) | | Descriptor: | RHODOPSIN | | Authors: | Yeagle, P.L, Salloum, A, Chopra, A, Bhawsar, N, Ali, L. | | Deposit date: | 2000-01-28 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the intradiskal loops and amino terminus of the G-protein receptor, rhodopsin.
J.Pept.Res., 55, 2000
|
|
1EDW
 
 | | SOLUTION STRUCTURE OF THIRD INTRADISKAL LOOP OF BOVINE RHODOPSIN (RESIDUES 268-293) | | Descriptor: | RHODOPSIN | | Authors: | Yeagle, P.L, Salloum, A, Chopra, A, Bhawsar, N, Ali, L. | | Deposit date: | 2000-01-28 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the intradiskal loops and amino terminus of the G-protein receptor, rhodopsin.
J.Pept.Res., 55, 2000
|
|
1EDX
 
 | | SOLUTION STRUCTURE OF AMINO TERMINUS OF BOVINE RHODOPSIN (RESIDUES 1-40) | | Descriptor: | RHODOPSIN | | Authors: | Yeagle, P.L, Salloum, A, Chopra, A, Bhawsar, N, Ali, L. | | Deposit date: | 2000-01-28 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the intradiskal loops and amino terminus of the G-protein receptor, rhodopsin.
J.Pept.Res., 55, 2000
|
|
1EDY
 
 | |
1EDZ
 
 | | STRUCTURE OF THE NAD-DEPENDENT 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE | | Authors: | Monzingo, A.F, Breksa, A, Ernst, S, Appling, D.R, Robertus, J.D. | | Deposit date: | 2000-01-28 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray structure of the NAD-dependent 5,10-methylenetetrahydrofolate dehydrogenase from Saccharomyces cerevisiae.
Protein Sci., 9, 2000
|
|
1EE0
 
 | | 2-PYRONE SYNTHASE COMPLEXED WITH ACETOACETYL-COA | | Descriptor: | 2-PYRONE SYNTHASE, ACETOACETYL-COENZYME A | | Authors: | Jez, J.M, Austin, M.B, Ferrer, J, Bowmann, M.E, Schroeder, J, Noel, J.P. | | Deposit date: | 2000-01-28 | | Release date: | 2001-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural control of polyketide formation in plant-specific polyketide synthases.
Chem.Biol., 7, 2000
|
|
1EE1
 
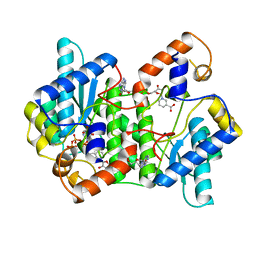 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH ONE MOLECULE ATP, TWO MOLECULES DEAMIDO-NAD+ AND ONE MG2+ ION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) SYNTHETASE, ... | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, Delucas, L. | | Deposit date: | 2000-01-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EE2
 
 | |
1EE3
 
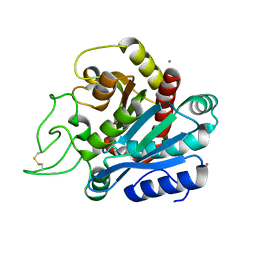 | | Cadmium-substituted bovine pancreatic carboxypeptidase A (alfa-form) at pH 7.5 and 2 mM chloride in monoclinic crystal form | | Descriptor: | CADMIUM ION, PROTEIN (CARBOXYPEPTIDASE A) | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-01-30 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
1EE4
 
 | |
1EE5
 
 | |
1EE6
 
 | | CRYSTAL STRUCTURE OF PECTATE LYASE FROM BACILLUS SP. STRAIN KSM-P15. | | Descriptor: | CALCIUM ION, PECTATE LYASE | | Authors: | Akita, M, Suzuki, A, Kobayashi, T, Ito, S, Yamane, T. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The first structure of pectate lyase belonging to polysaccharide lyase family 3.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EE7
 
 | | NMR STRUCTURE OF THE PEPTAIBOL CHRYSOSPERMIN C BOUND TO DPC MICELLES | | Descriptor: | CHRYSOSPERMIN C | | Authors: | Anders, R, Ohlenschlager, O, Soskic, V, Wenschuh, H, Heise, B, Brown, L.R. | | Deposit date: | 2000-01-31 | | Release date: | 2000-05-10 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The NMR Solution Structure of the Ion Channel Peptaibol Chrysospermin C Bound to Dodecylphosphocholine Micelles.
Eur.J.Biochem., 267, 2000
|
|
1EE8
 
 | | CRYSTAL STRUCTURE OF MUTM (FPG) PROTEIN FROM THERMUS THERMOPHILUS HB8 | | Descriptor: | MUTM (FPG) PROTEIN, ZINC ION | | Authors: | Sugahara, M, Mikawa, T, Kumasaka, T, Yamamoto, M, Kato, R, Fukuyama, K, Inoue, Y, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8.
EMBO J., 19, 2000
|
|
