6WSU
 
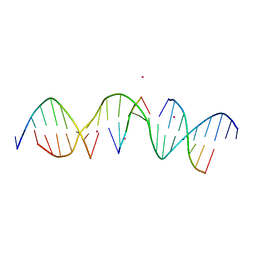 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J19 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*AP*GP*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSQ
 
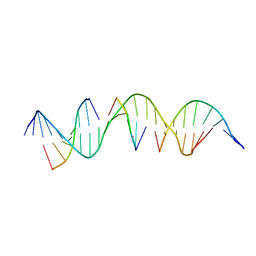 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J10 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSY
 
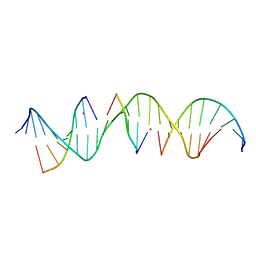 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J23 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*AP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.053 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSV
 
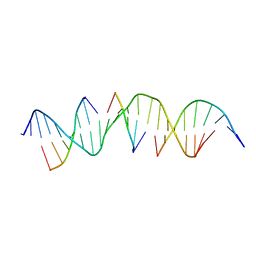 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J20 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*AP*CP*AP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WT0
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J25 immobile Holliday junction | | Descriptor: | COBALT (II) ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*AP*GP*AP*CP*CP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSS
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J15 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*AP*CP*CP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSP
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J9 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*AP*CP*TP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.049 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WT1
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J29 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*AP*GP*AP*CP*TP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), DNA (5'-D(P*AP*GP*TP*CP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSR
 
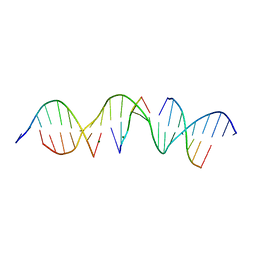 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J10 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*AP*GP*AP*CP*CP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSZ
 
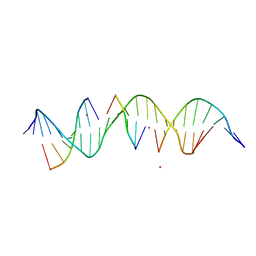 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J24 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.053 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSW
 
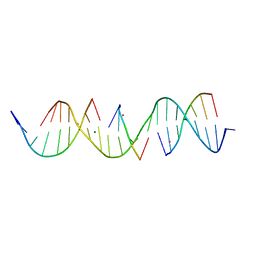 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J21 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*AP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*TP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSX
 
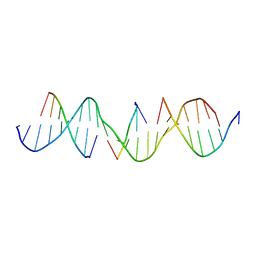 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J22 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.122 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
8D93
 
 | | [2T7] Self-assembling tensegrity triangle with R3 symmetry at 2.96 A resolution, update and junction cut for entry 3GBI | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*TP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*AP*CP*CP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Woloszyn, K, Lu, B, Sha, R, Ohayon, Y.P, Seeman, N.C. | | Deposit date: | 2022-06-09 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The Rule of Thirds: Controlling Junction Chirality and Polarity in 3D DNA Tiles.
Small, 19, 2023
|
|
1VQR
 
 | |
1EKW
 
 | | NMR STRUCTURE OF A DNA THREE-WAY JUNCTION | | Descriptor: | DNA (5'-D(*CP*GP*GP*TP*GP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*CP*AP*CP*CP*G)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*TP*CP*GP*CP*AP*GP*C)-3') | | Authors: | Thiviyanathan, V, Luxon, B.A, Leontis, N.B, Donne, D, Gorenstein, D.G. | | Deposit date: | 2000-03-09 | | Release date: | 2000-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hybrid-hybrid matrix structural refinement of a DNA three-way junction from 3D NOESY-NOESY.
J.Biomol.NMR, 14, 1999
|
|
1KBU
 
 | | CRE RECOMBINASE BOUND TO A LOXP HOLLIDAY JUNCTION | | Descriptor: | CRE RECOMBINASE, LOXP | | Authors: | Martin, S.S, Pulido, E, Chu, V.C, Lechner, T, Baldwin, E.P. | | Deposit date: | 2001-11-06 | | Release date: | 2002-06-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Order of Strand Exchanges in Cre-LoxP Recombination and its Basis Suggested by the Crystal Structure of a
Cre-LoxP Holliday Junction Complex
J.Mol.Biol., 319, 2002
|
|
1L4J
 
 | | Holliday Junction TCGGTACCGA with Na and Ca Binding Sites. | | Descriptor: | 5'-D(*TP*CP*GP*GP*TP*AP*CP*CP*GP*A)-3', CALCIUM ION, SODIUM ION | | Authors: | Thorpe, J.H, Gale, B.C, Teixeira, S.C.M, Cardin, C.J. | | Deposit date: | 2002-03-05 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational and Hydration Effects of Site-selective Sodium, Calcium and
Strontium Ion Binding to the DNA Holliday Junction Structure
d(TCGGTACCGA)4
J.Mol.Biol., 327, 2003
|
|
1KCF
 
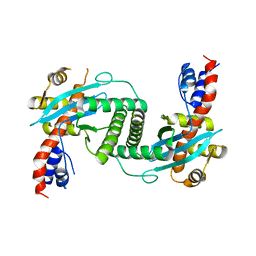 | | Crystal Structure of the Yeast Mitochondrial Holliday Junction Resolvase, Ydc2 | | Descriptor: | HYPOTHETICAL 30.2 KD PROTEIN C25G10.02 IN CHROMOSOME I, SULFATE ION | | Authors: | Ceschini, S, Keeley, A, McAlister, M.S.B, Oram, M, Phelan, J, Pearl, L.H, Tsaneva, I.R, Barrett, T.E. | | Deposit date: | 2001-11-08 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2.
EMBO J., 20, 2001
|
|
1IN8
 
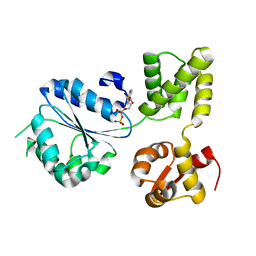 | | THERMOTOGA MARITIMA RUVB T158V | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1IN6
 
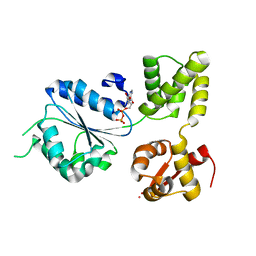 | | THERMOTOGA MARITIMA RUVB K64R MUTANT | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, ... | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1IN7
 
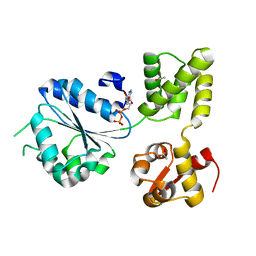 | | THERMOTOGA MARITIMA RUVB R170A | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1IN5
 
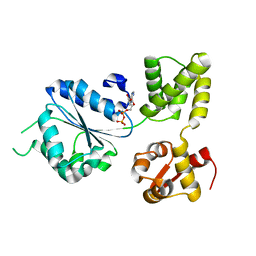 | | THERMOGOTA MARITIMA RUVB A156S MUTANT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Gonzalez, S, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1J7K
 
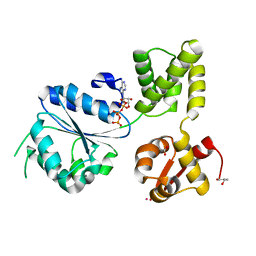 | | THERMOTOGA MARITIMA RUVB P216G MUTANT | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, COBALT (II) ION, ... | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-16 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1M6G
 
 | | Structural Characterisation of the Holliday Junction TCGGTACCGA | | Descriptor: | 5'-D(*TP*CP*GP*GP*TP*AP*CP*CP*GP*A)-3', STRONTIUM ION | | Authors: | Thorpe, J.H, Gale, B.C, Teixeira, S.C.M, Cardin, C.J. | | Deposit date: | 2002-07-16 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Conformational and hydration effects of site-selective sodium, calcium and
strontium ion binding to the DNA Holliday junction structure
d(TCGGTACCGA)(4)
J.Mol.Biol., 327, 2003
|
|
2QNC
 
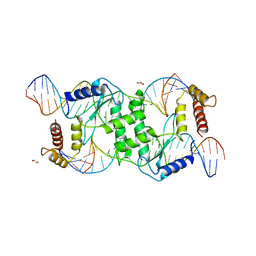 | | Crystal structure of T4 Endonuclease VII N62D mutant in complex with a DNA Holliday junction | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DAP*DGP*DGP*DCP*DCP*DTP*DAP*DGP*DCP*DGP*DTP*DCP*DCP*DGP*DGP*DAP*DAP*DTP*DTP*DCP*DTP*DTP*DCP*DG)-3'), DNA (5'-D(*DCP*DAP*DCP*DAP*DTP*DCP*DGP*DAP*DTP*DGP*DGP*DAP*DGP*DCP*DCP*DGP*DCP*DTP*DAP*DGP*DGP*DCP*DCP*DT)-3'), ... | | Authors: | Biertumpfel, C, Yang, W, Suck, D. | | Deposit date: | 2007-07-18 | | Release date: | 2008-01-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of T4 endonuclease VII resolving a Holliday junction.
Nature, 449, 2007
|
|
