2SBT
 
 | | A COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF SUBTILISIN BPN AND SUBTILISIN NOVO | | Descriptor: | ACETONE, SUBTILISIN NOVO | | Authors: | Drenth, J, Hol, W.G.J, Jansonius, J.N, Koekoek, R. | | Deposit date: | 1976-09-07 | | Release date: | 1976-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A comparison of the three-dimensional structures of subtilisin BPN' and subtilisin novo.
Cold Spring Harbor Symp.Quant.Biol., 36, 1972
|
|
3CNA
 
 | |
5PAD
 
 | |
4PAD
 
 | |
2PAD
 
 | |
6PAD
 
 | |
1PAD
 
 | |
2MHB
 
 | |
7LYZ
 
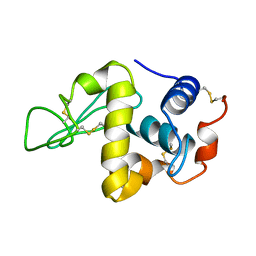 | | PROTEIN MODEL BUILDING BY THE USE OF A CONSTRAINED-RESTRAINED LEAST-SQUARES PROCEDURE | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Moult, J, Yonath, A, Sussman, J, Herzberg, O, Podjarny, A, Traub, W. | | Deposit date: | 1977-05-06 | | Release date: | 1977-06-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Model Building by the Use of a Constrained-Restrained Least-Squares Procedure
J.Appl.Crystallogr., 16, 1983
|
|
1PGI
 
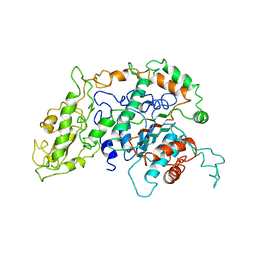 | |
8LYZ
 
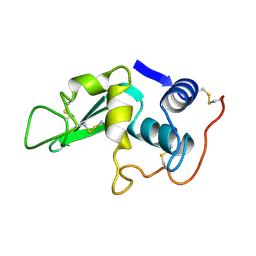 | |
2PAB
 
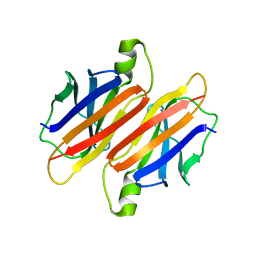 | |
1GCN
 
 | |
2HYA
 
 | | HYALURONIC ACID, MOLECULAR CONFORMATIONS AND INTERACTIONS IN TWO SODIUM SALTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid, SODIUM ION | | Authors: | Arnott, S. | | Deposit date: | 1977-11-20 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-21 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Hyaluronic acid: molecular conformations and interactions in two sodium salts.
J.Mol.Biol., 95, 1975
|
|
3HYA
 
 | | HYALURONIC ACID, MOLECULAR CONFORMATIONS AND INTERACTIONS IN TWO SODIUM SALTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid, SODIUM ION | | Authors: | Arnott, S. | | Deposit date: | 1977-11-20 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-21 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Hyaluronic acid: molecular conformations and interactions in two sodium salts.
J.Mol.Biol., 95, 1975
|
|
4HYA
 
 | | HYALURONIC ACID, THE ROLE OF DIVALENT CATIONS IN CONFORMATION AND PACKING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid, CALCIUM ION | | Authors: | Arnott, S. | | Deposit date: | 1977-11-20 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-28 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Hyaluronic acid: the role of divalent cations in conformation and packing.
J.Mol.Biol., 117, 1977
|
|
1HYA
 
 | |
1RHD
 
 | |
2YHX
 
 | |
4TNA
 
 | |
1CAP
 
 | | CONFORMATION AND MOLECULAR ORGANIZATION IN FIBERS OF THE CAPSULAR POLYSACCHARIDE FROM ESCHERICHIA COLI M41 MUTANT | | Descriptor: | alpha-D-mannopyranose-(1-3)-beta-D-glucopyranose-(1-3)-[4,6-O-[(1S)-1-carboxyethylidene]-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-4)]beta-D-glucopyranuronic acid-(1-3)-beta-D-galactopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-glucopyranose-(1-3)-[4,6-O-[(1S)-1-carboxyethylidene]-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-4)]beta-D-glucopyranuronic acid-(1-3)-beta-D-galactopyranose | | Authors: | Arnott, S. | | Deposit date: | 1978-05-23 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-07 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Conformation and molecular organization in fibers of the capsular polysaccharide from Escherichia coli M41 mutant.
J.Mol.Biol., 109, 1977
|
|
1C4S
 
 | | CHONDROITIN-4-SULFATE. THE STRUCTURE OF A SULFATED GLYCOSAMINOGLYCAN | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-4-deoxy-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-4-deoxy-beta-D-glucopyranuronic acid, SODIUM ION | | Authors: | Arnott, S. | | Deposit date: | 1978-05-23 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-07 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Chondroitin 4-sulfate: the structure of a sulfated glycosaminoglycan.
J.Mol.Biol., 125, 1978
|
|
1CAR
 
 | | I-CARRAGEENAN. MOLECULAR STRUCTURE AND PACKING OF POLYSACCHARIDE DOUBLE HELICES IN ORIENTED FIBRES OF DIVALENT CATION SALTS | | Descriptor: | 4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose | | Authors: | Arnott, S. | | Deposit date: | 1978-05-23 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-07 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Iota-carrageenan: molecular structure and packing of polysaccharide double helices in oriented fibres of divalent cation salts.
J.Mol.Biol., 90, 1974
|
|
1AGA
 
 | | THE AGAROSE DOUBLE HELIX AND ITS FUNCTION IN AGAROSE GEL STRUCTURE | | Descriptor: | beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose | | Authors: | Arnott, S. | | Deposit date: | 1978-05-23 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-07 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | The agarose double helix and its function in agarose gel structure.
J.Mol.Biol., 90, 1974
|
|
1KES
 
 | | CONFORMATION OF KERATAN SULPHATE | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose-(1-3)-6-O-sulfo-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose-(1-3)-6-O-sulfo-beta-D-galactopyranose | | Authors: | Arnott, S. | | Deposit date: | 1978-05-23 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-07 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Conformation of keratan sulphate.
J.Mol.Biol., 88, 1974
|
|
