5GM6
 
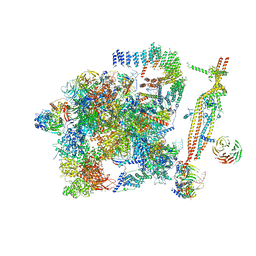 | | Cryo-EM structure of the activated spliceosome (Bact complex) at 3.5 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cold sensitive U2 snRNA suppressor 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yan, C, Wan, R, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-21 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a yeast activated spliceosome at 3.5 angstrom resolution
Science, 353, 2016
|
|
5FJ4
 
 | |
5EV4
 
 | |
5EV3
 
 | |
5EV2
 
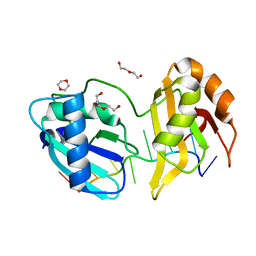 | | Structure II of Intact U2AF65 Recognizing the 3' Splice Site Signal | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DI(HYDROXYETHYL)ETHER, DNA (5'-R(P*UP*U)-D(P*UP*U)-R(P*U)-D(P*UP*(BRU)P*U)-3'), ... | | Authors: | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | An extended U2AF(65)-RNA-binding domain recognizes the 3' splice site signal.
Nat Commun, 7, 2016
|
|
5EV1
 
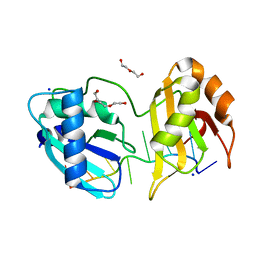 | | Structure I of Intact U2AF65 Recognizing a 3' Splice Site Signal | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA/RNA (5'-R(*UP*UP*U)-D(P*UP*UP*(BRU)P*U)-R(P*UP*U)-3'), SODIUM ION, ... | | Authors: | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.037 Å) | | Cite: | An extended U2AF(65)-RNA-binding domain recognizes the 3' splice site signal.
Nat Commun, 7, 2016
|
|
5EN1
 
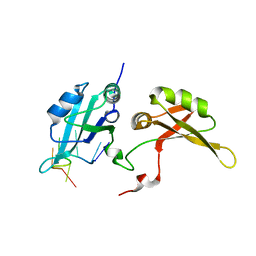 | | Crystal structure of hnRNPA2B1 in complex with RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*GP*GP*AP*CP*UP*G)-3') | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2015-11-09 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1
Nat Commun, 2018
|
|
5DET
 
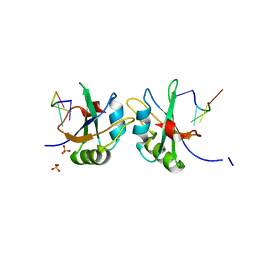 | | X-ray structure of human RBPMS in complex with the RNA | | Descriptor: | RNA (5'-R(*UP*CP*AP*C)-3'), RNA (5'-R(P*UP*CP*AP*CP*U)-3'), RNA-binding protein with multiple splicing, ... | | Authors: | Teplova, M, Farazi, T.A, Tuschl, T, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis underlying CAC RNA recognition by the RRM domain of dimeric RNA-binding protein RBPMS.
Q. Rev. Biophys., 49, 2016
|
|
5DDR
 
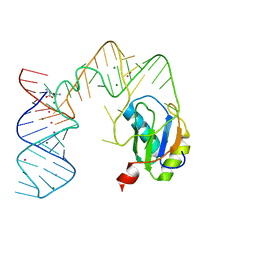 | | L-glutamine riboswitch bound with L-glutamine soaked with Cs+ | | Descriptor: | CESIUM ION, GLUTAMINE, L-glutamine riboswitch RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DDQ
 
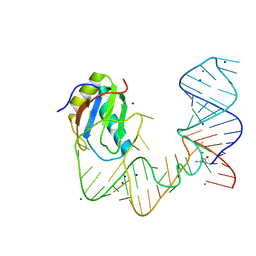 | | L-glutamine riboswitch bound with L-glutamine soaked with Mn2+ | | Descriptor: | GLUTAMINE, L-glutamine riboswitch RNA (61-MER), MAGNESIUM ION, ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DDP
 
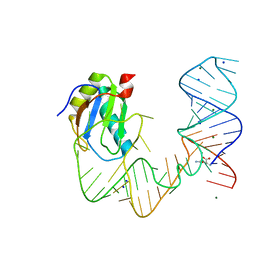 | | L-glutamine riboswitch bound with L-glutamine | | Descriptor: | GLUTAMINE, MAGNESIUM ION, RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DDO
 
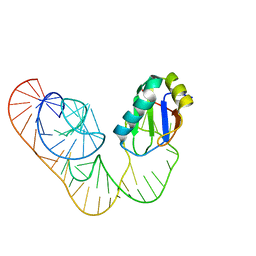 | |
5D78
 
 | |
5D77
 
 | | Structure of Mip6 RRM3 Domain | | Descriptor: | CITRIC ACID, NITRATE ION, RNA-binding protein MIP6, ... | | Authors: | Mohamad, N, Bravo, J. | | Deposit date: | 2015-08-13 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of Mip6 RRM3 domain at 1.3
To Be Published
|
|
5CYJ
 
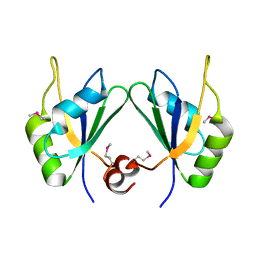 | | X-ray structure of human RBPMS | | Descriptor: | RNA-binding protein with multiple splicing | | Authors: | Teplova, M, Farazi, T.A, Patel, D.J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-09-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis underlying CAC RNA recognition by the RRM domain of dimeric RNA-binding protein RBPMS.
Q. Rev. Biophys., 49, 2016
|
|
5CA5
 
 | | Structure of the C. elegans NONO-1 homodimer | | Descriptor: | 1,2-ETHANEDIOL, NONO-1 | | Authors: | Knott, G.J, Bond, C.S. | | Deposit date: | 2015-06-29 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Caenorhabditis elegans NONO-1: Insights into DBHS protein structure, architecture, and function.
Protein Sci., 24, 2015
|
|
5BJR
 
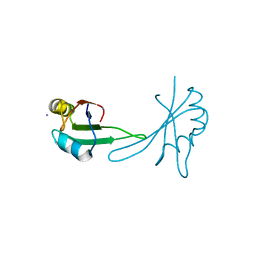 | |
5A5U
 
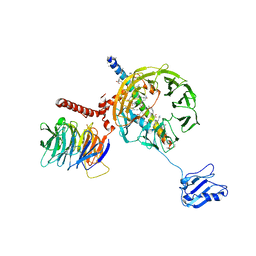 | | Structure of mammalian eIF3 in the context of the 43S preinitiation complex | | Descriptor: | EUKARYOTIC INITIATION FACTOR 3, EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SUBUNIT B, EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SUBUNIT I | | Authors: | des-Georges, A, Dhote, V, Kuhn, L, Hellen, C.U.T, Pestova, T.V, Frank, J, Hashem, Y. | | Deposit date: | 2015-06-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of Mammalian Eif3 in the Context of the 43S Preinitiation Complex.
Nature, 525, 2015
|
|
4ZKA
 
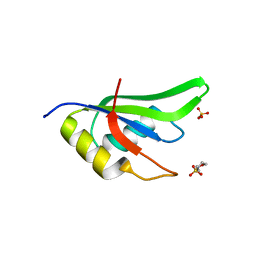 | | High Resolution Crystal Structure of Fox1 RRM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, RNA binding protein fox-1 homolog 1, SULFATE ION, ... | | Authors: | Blatter, M, Allain, F.H.-T. | | Deposit date: | 2015-04-30 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High Resolution Crystal Structure of Fox1 RRM
To Be Published
|
|
4Z2X
 
 | |
4YUD
 
 | |
4YOE
 
 | | Structure of UP1 bound to RNA 5'-AGU-3' | | Descriptor: | ACETATE ION, Heterogeneous nuclear ribonucleoprotein A1, RNA AGU, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2015-03-11 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The First Crystal Structure of the UP1 Domain of hnRNP A1 Bound to RNA Reveals a New Look for an Old RNA Binding Protein.
J.Mol.Biol., 427, 2015
|
|
4YH8
 
 | | Structure of yeast U2AF complex | | Descriptor: | Splicing factor U2AF 23 kDa subunit, Splicing factor U2AF 59 kDa subunit, ZINC ION | | Authors: | Yoshida, H, Park, S.Y, Urano, T, Obayashi, E. | | Deposit date: | 2015-02-27 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel 3' splice site recognition by the two zinc fingers in the U2AF small subunit.
Genes Dev., 29, 2015
|
|
4YB1
 
 | | 20A Mutant c-di-GMP Vc2 Riboswitch bound with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, MAGNESIUM ION, RNA (91-MER), ... | | Authors: | Ren, A.M, Patel, D.J, Rajashankar, R.K. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Structural Basis for Molecular Discrimination by a 3',3'-cGAMP Sensing Riboswitch.
Cell Rep, 11, 2015
|
|
4Y0F
 
 | | Crystal Structure of Human TDP-43 RRM1 Domain in Complex with an Unmodified Single-stranded DNA | | Descriptor: | DNA (5'-D(*GP*TP*TP*GP*AP*GP*CP*GP*TP*T)-3'), TAR DNA-binding protein 43 | | Authors: | Chiang, C.H, Kuo, P.H, Doudeva, L.G, Wang, Y.T, Yuan, H.S. | | Deposit date: | 2015-02-06 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Structural analysis of disease-related TDP-43 D169G mutation: linking enhanced stability and caspase cleavage efficiency to protein accumulation
Sci Rep, 6, 2016
|
|
