2DS2
 
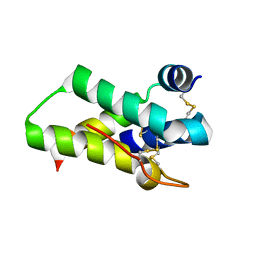 | | Crystal structure of mabinlin II | | Descriptor: | ACETIC ACID, Sweet protein mabinlin-2 chain A, Sweet protein mabinlin-2 chain B | | Authors: | Li, D.F, Zhu, D.Y, Wang, D.C. | | Deposit date: | 2006-06-19 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Mabinlin II: a novel structural type of sweet proteins and the main structural basis for its sweetness.
J.Struct.Biol., 162, 2008
|
|
4Z24
 
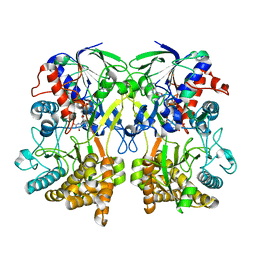 | | Mimivirus R135 (residues 51-702) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GMC-type oxidoreductase R135 | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2015-03-28 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mimivirus Enzyme that Participates in Viral Entry.
Structure, 23, 2015
|
|
4Z26
 
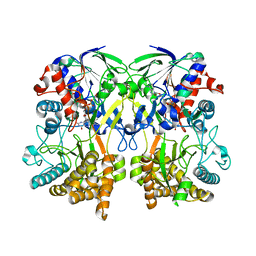 | | Mimivirus R135 (residues 51-702) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase R135 | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2015-03-28 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.915 Å) | | Cite: | A Mimivirus Enzyme that Participates in Viral Entry.
Structure, 23, 2015
|
|
2F52
 
 | |
5AGY
 
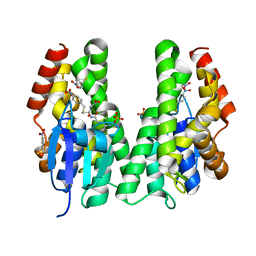 | | CRYSTAL STRUCTURE OF A TAU CLASS GST MUTANT FROM GLYCINE | | Descriptor: | 4-NITROPHENYL METHANETHIOL, GLUTATHIONE S-TRANSFERASE, PHOSPHATE ION, ... | | Authors: | Axarli, I, Muleta, A.W, Vlachakis, D, Kossida, S, Kotzia, G, Dhavala, P, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2015-02-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Directed Evolution of Tau Class Glutathione Transferases Reveals a Site that Regulates Catalytic Efficiency and Masks Cooperativity.
Biochem.J., 473, 2016
|
|
1Q6C
 
 | | Crystal Structure of Soybean Beta-Amylase Complexed with Maltose | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-amylase | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
1Q6D
 
 | | Crystal structure of Soybean Beta-Amylase Mutant (M51T) with Increased pH Optimum | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
1Q6F
 
 | | Crystal Structure of Soybean Beta-Amylase Mutant (E178Y) with Increased pH Optimum at pH 7.1 | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
8R44
 
 | | PAS-GAF bidomain of Glycine max phytochrome A | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, PHYCOCYANOBILIN, ... | | Authors: | Guan, K, Nagano, S, Hughes, J. | | Deposit date: | 2023-11-13 | | Release date: | 2025-01-08 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Pr and Pfr structures of plant phytochrome A.
Nat Commun, 16, 2025
|
|
8R45
 
 | | Phytochromobilin-adducted PAS-GAF bidomain of Glycine max phytochrome A | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A-2, TETRAETHYLENE GLYCOL | | Authors: | Guan, K, Chen, P, Nagano, S, Hughes, J. | | Deposit date: | 2023-11-13 | | Release date: | 2025-01-08 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pr and Pfr structures of plant phytochrome A.
Nat Commun, 16, 2025
|
|
3VBE
 
 | | Crystal structure of beta-cyanoalanine synthase in soybean | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, beta-cyanoalnine synthase | | Authors: | Yi, H, Jez, J.M. | | Deposit date: | 2012-01-02 | | Release date: | 2012-09-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Soybean beta-Cyanoalanine Synthase and the Molecular Basis for Cyanide Detoxification in Plants.
Plant Cell, 24, 2012
|
|
3VC3
 
 | | Crystal structure of beta-cyanoalanine synthase K95A mutant in soybean | | Descriptor: | N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE, beta-cyanoalnine synthase | | Authors: | Yi, H, Jez, J.M. | | Deposit date: | 2012-01-03 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.766 Å) | | Cite: | Structure of Soybean beta-Cyanoalanine Synthase and the Molecular Basis for Cyanide Detoxification in Plants.
Plant Cell, 24, 2012
|
|
4TVF
 
 | | OxyB from Actinoplanes teichomyceticus | | Descriptor: | OxyB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Haslinger, K, Cryle, M.J. | | Deposit date: | 2014-06-26 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cytochrome P450 OxyBtei Catalyzes the First Phenolic Coupling Step in Teicoplanin Biosynthesis.
Chembiochem, 15, 2014
|
|
7R6X
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2E12 Fab, S309 Fab, and S304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Monoclonal antibody S2E12 Fab heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
9MGN
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and compound 41 | | Descriptor: | 1,2-ETHANEDIOL, 2-(cyclobutylamino)-N-[(2S)-2-hydroxy-3-{6-[(1H-pyrazol-4-yl)methoxy]-3,4-dihydroisoquinolin-2(1H)-yl}propyl]pyridine-4-carboxamide, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2024-12-11 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | MTA-Cooperative PRMT5 Inhibitors: Mechanism Switching Through Structure-Based Design.
J.Med.Chem., 68, 2025
|
|
9MGR
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and compound 51 | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, 6-[(1-acetylazetidin-3-yl)amino]-N-[(2R)-2-hydroxy-2-{(3S)-7-[(4-methyl-1,3-oxazol-5-yl)methoxy]-1,2,3,4-tetrahydroisoquinolin-3-yl}ethyl]-2-(4-methylpiperidin-1-yl)pyrimidine-4-carboxamide, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2024-12-11 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | MTA-Cooperative PRMT5 Inhibitors: Mechanism Switching Through Structure-Based Design.
J.Med.Chem., 68, 2025
|
|
7R6W
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2X35 Fab and S309 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Snell, G, Czudnochowski, N, Hernandez, P, Nix, J.C, Croll, T.I, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
6LH8
 
 | | Structure of aerolysin-like protein (Bombina maxima) | | Descriptor: | aerolysin-like protein | | Authors: | Bian, X.L, Wang, Q.Q, Li, X, Teng, M.Q, Zhang, Y. | | Deposit date: | 2019-12-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | A cellular endolysosome-modulating pore-forming protein from a toad is negatively regulated by its paralog under oxidizing conditions.
J.Biol.Chem., 295, 2020
|
|
6LHZ
 
 | | Structure of aerolysin-like protein (Bombina maxima) | | Descriptor: | aerolysin-like protein | | Authors: | Bian, X.L, Wang, Q.Q, Li, X, Teng, M.Q, Zhang, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A cellular endolysosome-modulating pore-forming protein from a toad is negatively regulated by its paralog under oxidizing conditions.
J.Biol.Chem., 295, 2020
|
|
9N3R
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and TNG462 | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2025-01-31 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery of TNG462: A Highly Potent and Selective MTA-Cooperative PRMT5 Inhibitor to Target Cancers with MTAP Deletion.
J.Med.Chem., 68, 2025
|
|
9N3N
 
 | |
9N3O
 
 | |
9N3Q
 
 | |
9MGM
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and compound 24 | | Descriptor: | 2-(cyclobutylamino)-N-[(2S)-2-hydroxy-3-{6-[(1-methyl-1H-pyrazol-5-yl)methoxy]-3,4-dihydroisoquinolin-2(1H)-yl}propyl]pyridine-4-carboxamide, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2024-12-11 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | MTA-Cooperative PRMT5 Inhibitors: Mechanism Switching Through Structure-Based Design.
J.Med.Chem., 68, 2025
|
|
9N3P
 
 | |
