2DC3
 
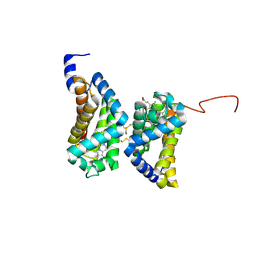 | | Crystal structure of human cytoglobin at 1.68 angstroms resolution | | Descriptor: | ACETIC ACID, Cytoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Makino, M, Sugimoto, H, Sawai, H, Kawada, N, Yoshizato, K, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-21 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | High-resolution structure of human cytoglobin: identification of extra N- and C-termini and a new dimerization mode.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2DC4
 
 | |
2DC5
 
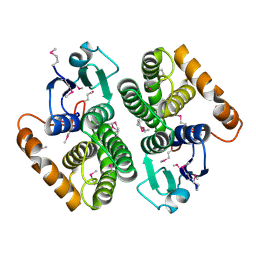 | | Crystal structure of mouse glutathione S-transferase, mu7 (GSTM7) at 1.6 A resolution | | Descriptor: | Glutathione S-transferase, mu 7 | | Authors: | Kamo, S, Kishishita, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-28 | | Release date: | 2006-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of mouse glutathione S-transferase, mu7 (GSTM7) at 1.6 A resolution
To be Published
|
|
2DC6
 
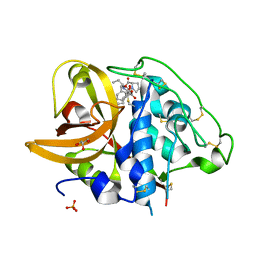 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA073 complex | | Descriptor: | BENZYL N-({(2S,3S)-3-[(PROPYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-28 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DC7
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA042 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-THREONYL-L-ISOLEUCINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-31 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DC8
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA059 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-ISOLEUCINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-31 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DC9
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA074Me complex | | Descriptor: | CATHEPSIN B, GLYCEROL, METHYL N-({(2S,3S)-3-[(PROPYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINATE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-31 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DCA
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA075 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-ISOLEUCYL-L-ALANINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DCB
 
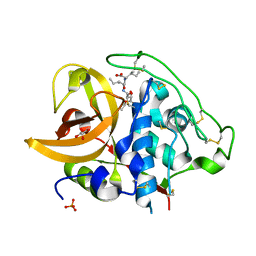 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA076 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-ISOLEUCYL-L-ISOLEUCINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DCC
 
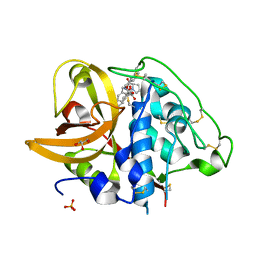 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA077 complex | | Descriptor: | BENZYL N-({(2S,3S)-3-[(BENZYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINATE, CATHEPSIN B, GLYCEROL, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DCD
 
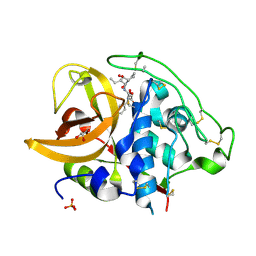 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA078 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-({(2S,3S)-3-[(BENZYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DCE
 
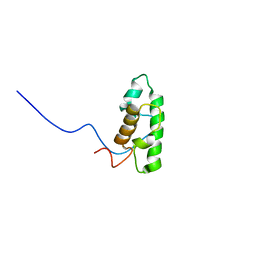 | | Solution structure of the SWIRM domain of human KIAA1915 protein | | Descriptor: | KIAA1915 protein | | Authors: | Yoneyama, M, Tochio, N, Umehara, T, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-06 | | Release date: | 2006-07-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Differences of SWIRM Domain Subtypes
J.Mol.Biol., 369, 2007
|
|
2DCF
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/H266N mutant with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2006-01-06 | | Release date: | 2007-01-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Nylon-oligomer degrading enzyme/substrate complex: catalytic mechanism of 6-aminohexanoate-dimer hydrolase
J.Mol.Biol., 370, 2007
|
|
2DCG
 
 | | MOLECULAR STRUCTURE OF A LEFT-HANDED DOUBLE HELICAL DNA FRAGMENT AT ATOMIC RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, SPERMINE | | Authors: | Wang, A.H.-J, Quigley, G.J, Kolpak, F.J, Crawford, J.L, Van Boom, J.H, Van Der Marel, G.A, Rich, A. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Molecular structure of a left-handed double helical DNA fragment at atomic resolution.
Nature, 282, 1979
|
|
2DCH
 
 | | Crystal structure of archaeal intron-encoded homing endonuclease I-Tsp061I | | Descriptor: | CHLORIDE ION, SULFATE ION, putative homing endonuclease | | Authors: | Nakayama, H, Tsuge, H, Shimamura, T, Miyano, M, Nomura, N, Sako, Y. | | Deposit date: | 2006-01-06 | | Release date: | 2006-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of a hyperthermophilic archaeal homing endonuclease, I-Tsp061I: contribution of cross-domain polar networks to thermostability.
J.Mol.Biol., 365, 2007
|
|
2DCI
 
 | |
2DCJ
 
 | | A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Ihsanawati, Tanaka, N, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-01-07 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1
To be Published
|
|
2DCK
 
 | | A tetragonal-lattice structure of alkaliphilic XynJ from Bacillus sp. 41M-1 | | Descriptor: | CALCIUM ION, GLYCEROL, xylanase J | | Authors: | Fibriansah, G, Ihsanawati, Tanaka, N, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-01-07 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1
To be Published
|
|
2DCL
 
 | |
2DCM
 
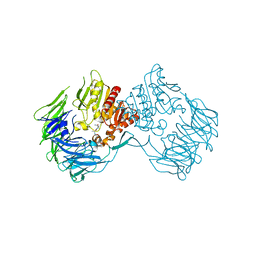 | | The Crystal Structure of S603A Mutated Prolyl Tripeptidyl Aminopeptidase Complexed with Substrate | | Descriptor: | GLYCYLALANYL-N-2-NAPHTHYL-L-PROLINEAMIDE, dipeptidyl aminopeptidase IV, putative | | Authors: | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | Deposit date: | 2006-01-09 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
2DCN
 
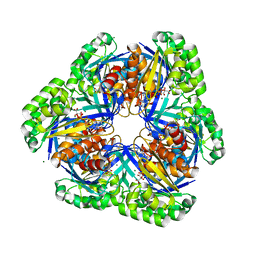 | | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate (alpha-furanose form) | | Descriptor: | 6-O-phosphono-beta-D-psicofuranosonic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Okazaki, S, Onda, H, Suzuki, A, Kuramitsu, S, Masui, R, Yamane, T. | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate
To be Published
|
|
2DCO
 
 | |
2DCP
 
 | |
2DCQ
 
 | |
2DCR
 
 | |
