2M51
 
 | |
2P0D
 
 | | ArhGAP9 PH domain in complex with Ins(1,4,5)P3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Rho GTPase-activating protein 9 | | Authors: | Ceccarelli, D.F.J, Blasutig, I, Goudreault, M, Ruston, J, Pawson, T, Sicheri, F. | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Non-canonical Interaction of Phosphoinositides with Pleckstrin Homology Domains of Tiam1 and ArhGAP9.
J.Biol.Chem., 282, 2007
|
|
2EXF
 
 | |
2OZM
 
 | | Crystal structure of RB69 gp43 in complex with DNA with 5-NITP opposite an abasic site analog | | Descriptor: | 1-{2-DEOXY-5-O-[(R)-HYDROXY{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}PHOSPHORYL]-BETA-D-ERYTHRO-PENTOFURANOSYL}-5-NITRO -1H-INDOLE, DNA polymerase, MAGNESIUM ION, ... | | Authors: | Zahn, K.E, Belrhali, H, Wallace, S.S, Doublie, S. | | Deposit date: | 2007-02-26 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Caught Bending the A-Rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide
Biochemistry, 46, 2007
|
|
2P27
 
 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with Mg2+ at 1.9 A resolution | | Descriptor: | MAGNESIUM ION, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2MF8
 
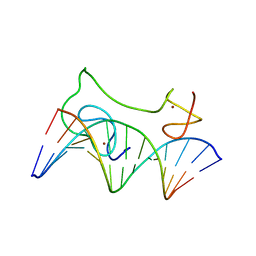 | | HADDOCK model of MyT1 F4F5 - DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*AP*AP*AP*GP*TP*TP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*AP*AP*CP*TP*TP*TP*CP*GP*GP*T)-3'), Myelin transcription factor 1, ... | | Authors: | Gamsjaeger, R, O'Connell, M.R, Cubeddu, L, Shepherd, N.E, Lowry, J.A, Kwan, A.H, Vandevenne, M, Swanton, M.K, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A structural analysis of DNA binding by myelin transcription factor 1 double zinc fingers.
J.Biol.Chem., 288, 2013
|
|
2PA4
 
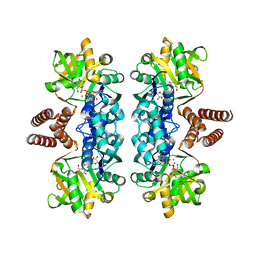 | |
2PTZ
 
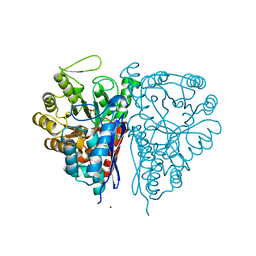 | | Crystal Structure of the T. brucei enolase complexed with phosphonoacetohydroxamate (PAH), His156-out conformation | | Descriptor: | 1,2-ETHANEDIOL, Enolase, PHOSPHONOACETOHYDROXAMIC ACID, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
3R6C
 
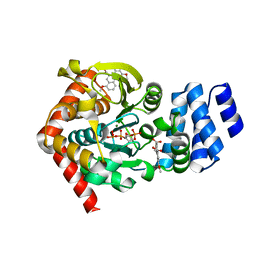 | | Anthranilate phosphoribosyltransferase (trpD) from Mycobacterium tuberculosis (complex with inhibitor ACS179) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 8-methoxyphenanthro[3,4-d][1,3]dioxole-5,6-dicarboxylic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Castell, A, Short, F.L, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-03-21 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The Substrate Capture Mechanism of Mycobacterium tuberculosis Anthranilate Phosphoribosyltransferase Provides a Mode for Inhibition.
Biochemistry, 52, 2013
|
|
3F9T
 
 | |
4GA9
 
 | |
1ETB
 
 | | THE X-RAY CRYSTAL STRUCTURE REFINEMENTS OF NORMAL HUMAN TRANSTHYRETIN AND THE AMYLOIDOGENIC VAL 30-->MET VARIANT TO 1.7 ANGSTROMS RESOLUTION | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, TRANSTHYRETIN | | Authors: | Braden, B.C, Steinrauf, L.K, Hamilton, J.A. | | Deposit date: | 1993-05-12 | | Release date: | 1995-01-26 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The x-ray crystal structure refinements of normal human transthyretin and the amyloidogenic Val-30-->Met variant to 1.7-A resolution.
J.Biol.Chem., 268, 1993
|
|
2QWK
 
 | |
4OUE
 
 | |
1REM
 
 | | HUMAN LYSOZYME WITH MAN-B1,4-GLCNAC COVALENTLY ATTACHED TO ASP53 | | Descriptor: | LYSOZYME, NITRATE ION, R-1,2-PROPANEDIOL, ... | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1998-01-14 | | Release date: | 1998-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of human lysozyme labelled with 2',3'-epoxypropyl beta-glycoside of man-beta1,4-GlcNAc. Structural change and recognition specificity at subsite B.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1ONW
 
 | | Crystal structure of Isoaspartyl Dipeptidase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Isoaspartyl dipeptidase, ... | | Authors: | Thoden, J.B, Marti-Arbona, R, Raushel, F.M, Holden, H.M. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-06 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High Resolution X-ray Structure of
Isoaspartyl Dipeptidase from
Escherichia coli
Biochemistry, 42, 2003
|
|
4FW7
 
 | |
3NP9
 
 | |
2KB5
 
 | | Solution NMR Structure of Eosinophil Cationic Protein/RNase 3 | | Descriptor: | Eosinophil cationic protein | | Authors: | Rico, M, Bruix, M, Laurents, D.V, Santoro, J, Jimenez, M, Boix, E, Moussaoui, M, Nogues, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-06-23 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The (1)H, (13)C, (15)N resonance assignment, solution structure, and residue level stability of eosinophil cationic protein/RNase 3 determined by NMR spectroscopy
Biopolymers, 91, 2009
|
|
1ET1
 
 | | CRYSTAL STRUCTURE OF HUMAN PARATHYROID HORMONE 1-34 AT 0.9 A RESOLUTION | | Descriptor: | PARATHYROID HORMONE, SODIUM ION | | Authors: | Jin, L, Briggs, S.L, Chandrasekhar, S, Chirgadze, N.Y, Clawson, D.K, Schevitz, R.W, Smiley, D.L, Tashjian, A.H, Zhang, F. | | Deposit date: | 2000-04-12 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal structure of human parathyroid hormone 1-34 at 0.9-A resolution.
J.Biol.Chem., 275, 2000
|
|
3GXR
 
 | | The crystal structure of g-type lysozyme from Atlantic cod (Gadus morhua L.) in complex with NAG oligomers sheds new light on substrate binding and the catalytic mechanism. Structure with NAG to 1.7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Goose-type lysozyme 1 | | Authors: | Helland, R, Larsen, R.L, Finstad, S, Kyomuhendo, P, Larsen, A.N. | | Deposit date: | 2009-04-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of g-type lysozyme from Atlantic cod shed new light on substrate binding and the catalytic mechanism.
Cell.Mol.Life Sci., 66, 2009
|
|
1R4W
 
 | | Crystal structure of Mitochondrial class kappa glutathione transferase | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, mitochondrial | | Authors: | Ladner, J.E, Parsons, J.F, Rife, C.L, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2003-10-08 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Parallel Evolutionary Pathways for Glutathione Transferases: Structure and Mechanism of the Mitochondrial Class Kappa Enzyme rGSTK1-1
Biochemistry, 43, 2004
|
|
1R4P
 
 | | Shiga toxin type 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, FORMIC ACID, ... | | Authors: | Fraser, M.E, Fujinaga, M, Cherney, M.M, Melton-Celsa, A.R, Twiddy, E.M, O'Brien, A.D, James, M.N.G. | | Deposit date: | 2003-10-07 | | Release date: | 2004-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of Shiga Toxin Type 2 (Stx2) from Escherichia coli O157:H7.
J.Biol.Chem., 279, 2004
|
|
3O3U
 
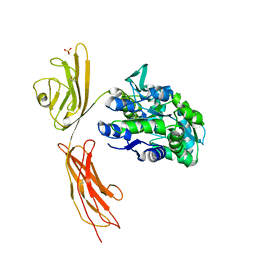 | | Crystal Structure of Human Receptor for Advanced Glycation Endproducts (RAGE) | | Descriptor: | Maltose-binding periplasmic protein, Advanced glycosylation end product-specific receptor, SULFATE ION, ... | | Authors: | Park, H, Boyington, J.C. | | Deposit date: | 2010-07-26 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | The 1.5 A crystal structure of human receptor for advanced glycation endproducts (RAGE) ectodomains reveals unique features determining ligand binding.
J.Biol.Chem., 285, 2010
|
|
2QQM
 
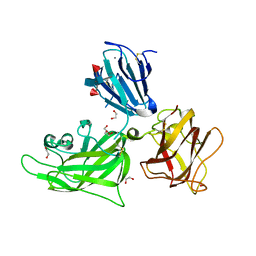 | | Crystal Structure of the a2b1b2 Domains from Human Neuropilin-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Appleton, B.A, Wiesmann, C. | | Deposit date: | 2007-07-26 | | Release date: | 2007-11-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of neuropilin/antibody complexes provide insights into semaphorin and VEGF binding
Embo J., 26, 2007
|
|
