1U6F
 
 | | NMR solution structure of TcUBP1, a single RBD-unit from Trypanosoma cruzi | | Descriptor: | RNA-binding protein UBP1 | | Authors: | Volpon, L, D'orso, I, Frasch, A, Gehring, K. | | Deposit date: | 2004-07-29 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Study of TcUBP1, a Single RRM Domain Protein from Trypanosoma cruzi: Contribution of a beta Hairpin to RNA Binding
Biochemistry, 44, 2005
|
|
2OFQ
 
 | | NMR Solution Structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system | | Descriptor: | TraN, TraO | | Authors: | Harris, R, Bayliss, R, Driscoll, P.C, Waksman, G. | | Deposit date: | 2007-01-04 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1T0V
 
 | | NMR Solution Structure of the Engineered Lipocalin FluA(R95K) Northeast Structural Genomics Target OR17 | | Descriptor: | BILIN-BINDING PROTEIN | | Authors: | Mills, J.L, Liu, G, Skerra, A, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-04-13 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of the engineered fluorescein-binding lipocalin FluA reveal rigidification of beta-barrel and variable loops upon enthalpy-driven ligand binding.
Biochemistry, 48, 2009
|
|
1WWN
 
 | | NMR Solution Structure of BmK-betaIT, an Excitatory Scorpion Toxin from Buthus martensi Karsch | | Descriptor: | Excitatory insect selective toxin 1 | | Authors: | Wu, H, Tong, X, Chen, X, Zhang, Q, Zheng, X, Zhang, N, Wu, G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of BmK-betaIT, an excitatory scorpion beta-toxin without a 'hot spot' at the relevant position
Biochem.Biophys.Res.Commun., 349, 2006
|
|
1X2U
 
 | |
1X2X
 
 | |
1X2V
 
 | |
1X2Y
 
 | |
1X30
 
 | |
2JPO
 
 | |
1XBL
 
 | | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | | Descriptor: | DNAJ | | Authors: | Pellecchia, M, Szyperski, T, Wall, D, Georgopoulos, C, Wuthrich, K. | | Deposit date: | 1996-10-07 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone.
J.Mol.Biol., 260, 1996
|
|
1Z66
 
 | | NMR solution structure of domain III of E-protein of tick-borne Langat flavivirus (no RDC restraints) | | Descriptor: | Major envelope protein E | | Authors: | Mukherjee, M, Dutta, K, White, M.A, Cowburn, D, Fox, R.O. | | Deposit date: | 2005-03-21 | | Release date: | 2006-03-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and backbone dynamics of domain III of the E protein of tick-borne Langat flavivirus suggests a potential site for molecular recognition.
Protein Sci., 15, 2006
|
|
2B3I
 
 | | NMR SOLUTION STRUCTURE OF PLASTOCYANIN FROM THE PHOTOSYNTHETIC PROKARYOTE, PROCHLOROTHRIX HOLLANDICA (19 STRUCTURES) | | Descriptor: | COPPER (I) ION, PROTEIN (PLASTOCYANIN) | | Authors: | Babu, C.R, Volkman, B.F, Bullerjahn, G.S. | | Deposit date: | 1998-12-11 | | Release date: | 1999-04-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of plastocyanin from the photosynthetic prokaryote, Prochlorothrix hollandica.
Biochemistry, 38, 1999
|
|
2B0F
 
 | |
2KXV
 
 | |
2KXT
 
 | |
2IN2
 
 | |
1YGM
 
 | | NMR structure of Mistic | | Descriptor: | hypothetical protein BSU31320 | | Authors: | Roosild, T.P, Greenwald, J, Vega, M, Castronovo, S, Riek, R, Choe, S. | | Deposit date: | 2005-01-05 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Mistic, a membrane-integrating protein for membrane protein expression.
Science, 307, 2005
|
|
2JSS
 
 | | NMR structure of chaperone Chz1 complexed with histone H2A.Z-H2B | | Descriptor: | Chimera of Histone H2B.1 and Histone H2A.Z, Uncharacterized protein YER030W | | Authors: | Zhou, Z, Feng, H, Hansen, D.F, Kato, H, Luk, E, Freedberg, D.I, Kay, L.E, Wu, C, Bai, Y. | | Deposit date: | 2007-07-11 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2K7L
 
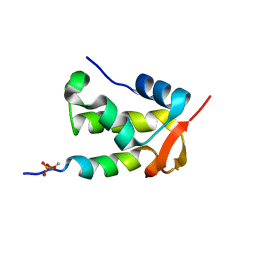 | | NMR structure of a complex formed by the C-terminal domain of human RAP74 and a phosphorylated peptide from the central domain of the FCP1 | | Descriptor: | General transcription factor IIF subunit 1, centFCP1-T584PO4 peptide | | Authors: | Yang, A, Abbott, K.L, Desjardins, A, Di Lello, P, Omichinski, J.G, Legault, P. | | Deposit date: | 2008-08-13 | | Release date: | 2009-06-02 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a complex formed by the carboxyl-terminal domain of human RAP74 and a phosphorylated peptide from the central domain of the FCP1 phosphatase
Biochemistry, 48, 2009
|
|
2M2I
 
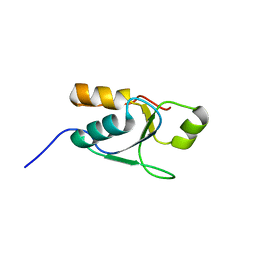 | |
1TNX
 
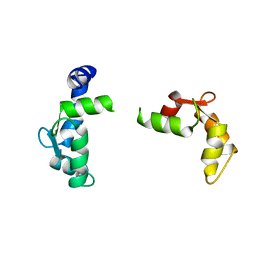 | |
1TNW
 
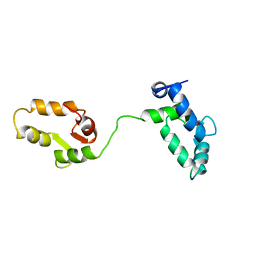 | |
1TRL
 
 | | NMR SOLUTION STRUCTURE OF THE C-TERMINAL FRAGMENT 255-316 OF THERMOLYSIN: A DIMER FORMED BY SUBUNITS HAVING THE NATIVE STRUCTURE | | Descriptor: | THERMOLYSIN FRAGMENT 255 - 316 | | Authors: | Rico, M, Jimenez, M.A, Gonzalez, C, De Filippis, V, Fontana, A. | | Deposit date: | 1994-09-02 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the C-terminal fragment 255-316 of thermolysin: a dimer formed by subunits having the native structure.
Biochemistry, 33, 1994
|
|
1T50
 
 | | NMR SOLUTION STRUCTURE OF APLYSIA ATTRACTIN | | Descriptor: | Attractin | | Authors: | Ravindranath, G, Xu, Y, Schein, C.H, Rajaratnam, K, Painter, S.D, Nagle, G.T, Braun, W. | | Deposit date: | 2004-04-30 | | Release date: | 2004-05-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Attractin, a Water-Borne Pheromone from the Mollusk Aplysia Attractin
Biochemistry, 42, 2003
|
|
