8FU9
 
 | | Structure of Covid Spike variant deltaN25 with one erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
6M7Z
 
 | | A divergent kinase lacking the glycine-rich loop regulates membrane ultrastructure of the Toxoplasma parasitophorous vacuole | | Descriptor: | 1,2-ETHANEDIOL, Bradyzoite pseudokinase 1, CHLORIDE ION | | Authors: | Beraki, T, Borek, D.M, Reese, M.L. | | Deposit date: | 2018-08-21 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Divergent kinase regulates membrane ultrastructure of theToxoplasmaparasitophorous vacuole.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6MT9
 
 | | X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP, ATP, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-19 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6MV9
 
 | | X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-24 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
4IRO
 
 | | Crystal structure of T-state carbonmonoxy hemoglobin from Trematomus bernacchii at pH 8.4 | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Merlino, A, Balsamo, A, Mazzarella, L, Vergara, A. | | Deposit date: | 2013-01-15 | | Release date: | 2013-02-20 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of tertiary structures on the Root effect in fish hemoglobins.
Biochim.Biophys.Acta, 1834, 2013
|
|
6MVE
 
 | | Reduced X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP, ATP, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-25 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
8GJC
 
 | | X-ray crystallographic structure of a beta-hairpin peptide derived from Abeta 17-35. (ORN)LVFFAED(ORN)GAI(N-Me-Ile)GLM | | Descriptor: | DI(HYDROXYETHYL)ETHER, METHIONINE, beta-hairpin peptide derived from Abeta 17-35, ... | | Authors: | Kreutzer, A.G, Ruttenberg, S.M, Nowick, J.S. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.431 Å) | | Cite: | beta-Hairpin Alignment Alters Oligomer Formation in A beta-Derived Peptides.
Biochemistry, 63, 2024
|
|
8GJD
 
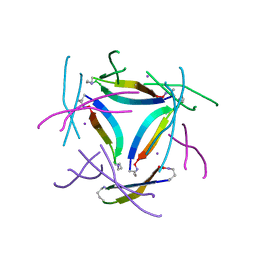 | | X-ray crystallographic structure of a beta-hairpin peptide derived from Abeta 17-36. (ORN)LVFFAED(ORN)AII(N-Me-Gly)LMV | | Descriptor: | Beta-hairpin peptide derived from Abeta 17-36, IODIDE ION | | Authors: | Kreutzer, A.G, Ruttenberg, S.M, Nowick, J.S, Truex, N.L. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | beta-Hairpin Alignment Alters Oligomer Formation in A beta-Derived Peptides.
Biochemistry, 63, 2024
|
|
6MYX
 
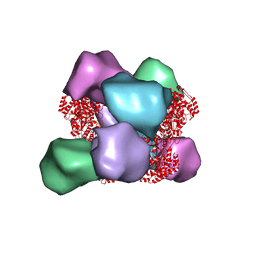 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited double-helical filament of NrdE alpha subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase | | Authors: | Thomas, W.C, Bacik, J.P, Chen, J.Z, Ando, N. | | Deposit date: | 2018-11-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
7FJH
 
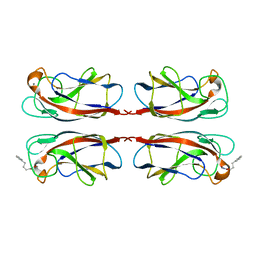 | | LecA from Pseudomonas aeruginosa in complex with 4-Phenylbutyryl hydroxamic acid (CAS: 32153-46-1) | | Descriptor: | CALCIUM ION, N-oxidanyl-4-phenyl-butanamide, PA-I galactophilic lectin | | Authors: | Shanina, S, Kuhaudomlarp, S, Siebs, E, Fuchsberger, F, Denis, M, da Silva Figueiredo Celstino Gomes, P, Clausen, M.H, Seeberger, P.H, Rognan, D, Titz, A, Imberty, A, Rademacher, C. | | Deposit date: | 2021-08-04 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Targeting undruggable carbohydrate recognition sites through focused fragment library design.
Commun Chem, 5, 2022
|
|
4JWQ
 
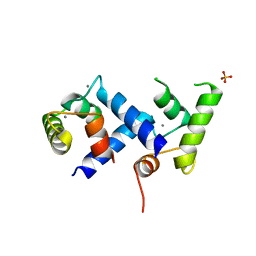 | | Crystal Structure of the Calcium Binding Domain of CDPK3 from Plasmodium Berghei, PB000947.00 | | Descriptor: | CALCIUM ION, Calcium-dependent protein kinase, SULFATE ION | | Authors: | Wernimont, A.K, Loppnau, P, Lin, Y.H, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Hui, R, Mottaghi, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-27 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Calcium Binding Domain of CDPK3 from Plasmodium Berghei, PB000947.00
TO BE PUBLISHED
|
|
8FU7
 
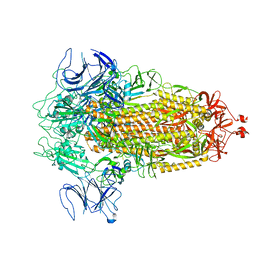 | | Structure of Covid Spike variant deltaN135 in fully closed form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
8FU8
 
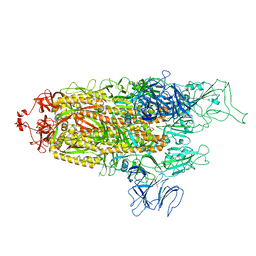 | | Structure of Covid Spike variant deltaN135 with one erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
6MW3
 
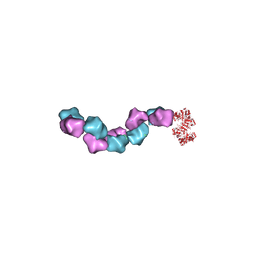 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited filament composed of NrdE alpha subunit and NrdF beta subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase, Ribonucleoside-diphosphate reductase NrdF beta subunit | | Authors: | Thomas, W.C, Bacik, J.P, Kaelber, J.T, Ando, N. | | Deposit date: | 2018-10-29 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
4N7C
 
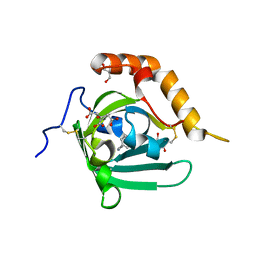 | | Structural re-examination of native Bla g 4 | | Descriptor: | 4-(2-aminoethyl)phenol, Bla g 4 allergen variant 1, CITRIC ACID, ... | | Authors: | Offermann, L.R, Chan, S.L, Osinski, T, Tan, Y.W, Chew, F.T, Sivaraman, J, Mok, Y.K, Minor, W, Chruszcz, M. | | Deposit date: | 2013-10-15 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The major cockroach allergen Bla g 4 binds tyramine and octopamine.
Mol.Immunol., 60, 2014
|
|
4L53
 
 | | Crystal Structure of (1R,4R)-4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)-3-chlorofuro[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}cyclohexan-1-ol bound to TAK1-TAB1 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera, ... | | Authors: | Wang, J, Hornberger, K.R, Crew, A.P, Jestel, A, Maskos, K, Moertl, M. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1: Optimization of kinase selectivity and pharmacokinetics.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5OTJ
 
 | | Monomeric polcalcin (Phl p 7) in complex with two identical allergen-specific antibodies | | Descriptor: | 1,2-ETHANEDIOL, 102.1F10 Fab heavy chain, 102.1F10 Fab light chain, ... | | Authors: | Mitropoulou, A.N, Davies, A.M, Beavil, A.J, McDonnell, J.M, Sutton, B.J. | | Deposit date: | 2017-08-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a patient-derived antibody in complex with allergen reveals simultaneous conventional and superantigen-like recognition.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4M3B
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-methyl-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
4L3N
 
 | | Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Chen, Y, Rajashankar, K.R, Yang, Y, Agnihothram, S.S, Liu, C, Lin, Y.-L, Baric, R.S, Li, F. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of the receptor-binding domain from newly emerged middle East respiratory syndrome coronavirus.
J.Virol., 87, 2013
|
|
4HW2
 
 | | Discovery of potent Mcl-1 inhibitors using fragment-based methods and structure-based design | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Zhao, B. | | Deposit date: | 2012-11-07 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of potent myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods and structure-based design.
J.Med.Chem., 56, 2013
|
|
4HW3
 
 | | Discovery of potent Mcl-1 inhibitors using fragment-based methods and structure-based design | | Descriptor: | 3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-benzothiophene-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2012-11-07 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods and structure-based design.
J.Med.Chem., 56, 2013
|
|
4LDU
 
 | | Crystal structure of the DNA binding domain of Arabidopsis thaliana auxin response factor 5 | | Descriptor: | Auxin response factor 5, CHLORIDE ION | | Authors: | Boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
4LDV
 
 | | Crystal structure of the DNA binding domain of A. thailana auxin response factor 1 | | Descriptor: | Auxin response factor 1, CHLORIDE ION, FORMIC ACID, ... | | Authors: | boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
4C5Z
 
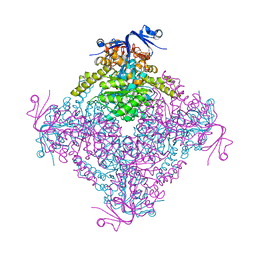 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
4IF5
 
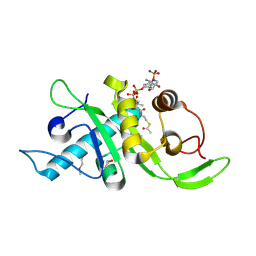 | | Structure of human Mec17 | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase, CHLORIDE ION | | Authors: | Davenport, A.M, Collins, L, Minor, P, Sternberg, P, Hoelz, A. | | Deposit date: | 2012-12-14 | | Release date: | 2014-05-28 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characterization of the alpha-Tubulin Acetyltransferase MEC-17.
J.Mol.Biol., 426, 2014
|
|
