1G4R
 
 | | CRYSTAL STRUCTURE OF BOVINE BETA-ARRESTIN 1 | | Descriptor: | BETA-ARRESTIN 1 | | Authors: | Schubert, C, Han, M. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation.
Structure, 9, 2001
|
|
4CLY
 
 | | Crystal structure of human soluble Adenylyl Cyclase soaked with biselenite | | Descriptor: | 1,2-ETHANEDIOL, ADENYLATE CYCLASE TYPE 10, BISELENITE ION, ... | | Authors: | Kleinboelting, S, Weyand, M, Steegborn, C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structures of Human Soluble Adenylyl Cyclase Reveal Mechanisms of Catalysis and of its Activation Through Bicarbonate.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CMX
 
 | | Crystal structure of Rv3378c | | Descriptor: | 1,2-ETHANEDIOL, BENZENE HEXACARBOXYLIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Layre, E, Lee, H.J, Young, D.C, Martinot, A.J, Buter, J, Minnaard, A.J, Annand, J.W, Fortune, S.M, Snider, B.B, Matsunaga, I, Rubin, E.J, Alber, T, Moody, D.B. | | Deposit date: | 2014-01-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular Profiling of Mycobacterium Tuberculosis Identifies Tuberculosinyl Nucleoside Products of the Virulence-Associated Enzyme Rv3378C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1GIA
 
 | |
3WK4
 
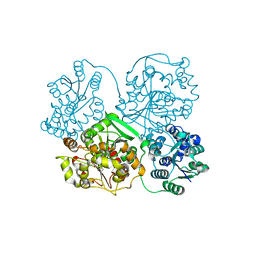 | | Crystal structure of soluble epoxide hydrolase in complex with fragment inhibitor | | Descriptor: | 1-[(1R)-1-cyclopropylethyl]-3-phenylurea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Amano, Y, Yamaguchi, T, Tanabe, E. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insights into binding of inhibitors to soluble epoxide hydrolase gained by fragment screening and X-ray crystallography.
Bioorg.Med.Chem., 22, 2014
|
|
7JID
 
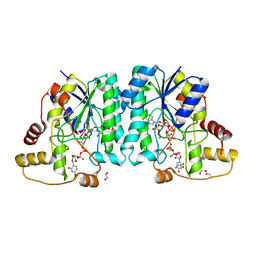 | | Crystal structure of the L780 UDP-rhamnose synthase from Acanthamoeba polyphaga mimivirus | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-L-Rhamnose Synthase, ... | | Authors: | Bockhaus, N.J, Ferek, J.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The high-resolution structure of a UDP-L-rhamnose synthase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 29, 2020
|
|
4PS7
 
 | |
5EQY
 
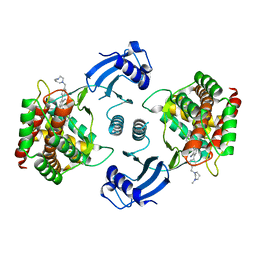 | | Crystal structure of choline kinase alpha-1 bound by 5-[(4-methyl-1,4-diazepan-1-yl)methyl]-2-[4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]benzenecarbonitrile (compound 65) | | Descriptor: | 5-[(4-methyl-1,4-diazepan-1-yl)methyl]-2-[4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]benzenecarbonitrile, Choline kinase alpha | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel Small Molecule Inhibitors of Choline Kinase Identified by Fragment-Based Drug Discovery.
J.Med.Chem., 59, 2016
|
|
2NSL
 
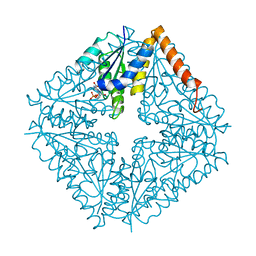 | | E. coli PurE H45N mutant complexed with CAIR | | Descriptor: | 5-AMINO-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-11-04 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N(5)-CAIR Mutase: Role of a CO(2) Binding Site and Substrate Movement in Catalysis.
Biochemistry, 46, 2007
|
|
3CCF
 
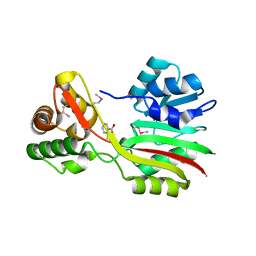 | |
5HO9
 
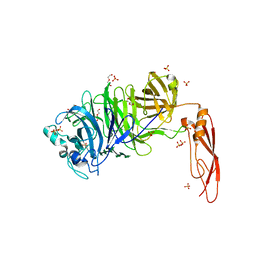 | | Structure of truncated AbnA (domains 1-3), a GH43 arabinanase from Geobacilllus stearothermophilus, in complex with arabinooctaose | | Descriptor: | DI(HYDROXYETHYL)ETHER, Extracellular arabinanase, SULFATE ION, ... | | Authors: | Lansky, S, Salama, R, Azoulai, D, Shwartstien, O, Shoham, Y, Shoham, G. | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | Structure of truncated AbnA (domains 1-3), a GH43 arabinanase from Geobacilllus stearothermophilus, in complex with arabinooctaose
To Be Published
|
|
6CQV
 
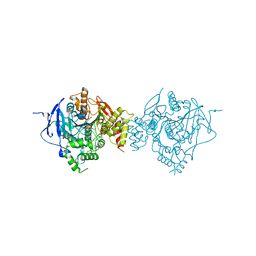 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with VX(+) and HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bester, S.M, Guelta, M.A, Pegan, S.D, Height, J.J. | | Deposit date: | 2018-03-16 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural Insights of Stereospecific Inhibition of Human Acetylcholinesterase by VX and Subsequent Reactivation by HI-6.
Chem. Res. Toxicol., 31, 2018
|
|
7JNR
 
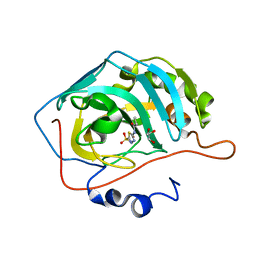 | | Carbonic Anhydrase II Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propionamide | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, ... | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.443 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
7JO2
 
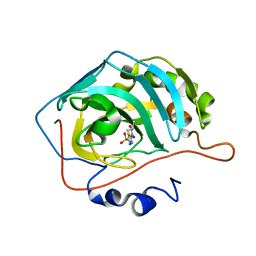 | | Carbonic Anhydrase IX Mimic Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)isobutyramide | | Descriptor: | 2-methyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.307 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
3BMN
 
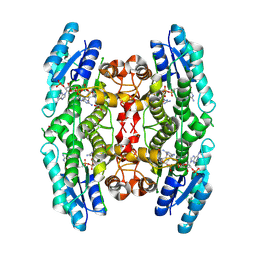 | | Structure of Pteridine Reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor (Compound AX3) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Martini, V.P, Iulek, J, Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2007-12-13 | | Release date: | 2008-12-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3WY9
 
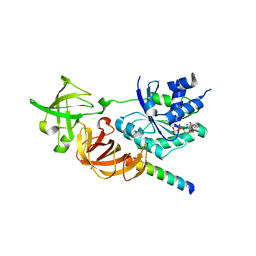 | | Crystal structure of a complex of the archaeal ribosomal stalk protein aP1 and the GDP-bound archaeal elongation factor aEF1alpha | | Descriptor: | 50S ribosomal protein L12, Elongation factor 1-alpha, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ito, K, Honda, T, Suzuki, T, Miyoshi, T, Murakami, R, Yao, M, Uchiumi, T. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular insights into the interaction of the ribosomal stalk protein with elongation factor 1 alpha.
Nucleic Acids Res., 42, 2014
|
|
1S6B
 
 | | X-ray Crystal Structure of a Complex Formed Between Two Homologous Isoforms of Phospholipase A2 from Naja naja sagittifera: Principle of Molecular Association and Inactivation | | Descriptor: | ACETIC ACID, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Jabeen, T, Sharma, S, Singh, R.K, Kaur, P, Singh, T.P. | | Deposit date: | 2004-01-23 | | Release date: | 2004-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a calcium-induced dimer of two isoforms of cobra phospholipase A2 at 1.6 A resolution.
Proteins, 59, 2005
|
|
2WK9
 
 | | Structure of Plp_Thr aldimine form of Vibrio cholerae CqsA | | Descriptor: | CAI-1 AUTOINDUCER SYNTHASE, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Jahan, N, Potter, J.A, Sheikh, M.A, Botting, C.H, Shirran, S.L, Westwood, N.J, Taylor, G.L. | | Deposit date: | 2009-06-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights Into the Biosynthesis of the Vibrio Cholerae Major Autoinducer Cai-1 from the Crystal Structure of the Plp-Dependent Enzyme Cqsa.
J.Mol.Biol., 392, 2009
|
|
3LRP
 
 | |
1WU1
 
 | |
4L8P
 
 | |
2WY0
 
 | | Crystal structure of mouse angiotensinogen in the oxidised form with space group P6122 | | Descriptor: | 1,2-ETHANEDIOL, ANGIOTENSINOGEN, SODIUM ION | | Authors: | Zhou, A, Wei, Z, Carrell, R.W, Read, R.J. | | Deposit date: | 2009-11-11 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A Redox Switch in Angiotensinogen Modulates Angiotensin Release.
Nature, 468, 2010
|
|
4D6D
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the blood group A-trisaccharide (X02 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
7E9L
 
 | | Crystal Structure of POMGNT2 in complex with UDP and mono-mannosyl peptide (379Man short peptide) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2021-03-04 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of POMGNT2 provides new insights into the mechanism to determine the functional O-mannosylation site on alpha-dystroglycan.
Genes Cells, 26, 2021
|
|
7E9K
 
 | | Crystal Structure of POMGNT2 in complex with UDP and mono-mannosyl peptide (379Man long peptide) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2021-03-04 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of POMGNT2 provides new insights into the mechanism to determine the functional O-mannosylation site on alpha-dystroglycan.
Genes Cells, 26, 2021
|
|
