9LO2
 
 | | Crystal Structure of Anti-CRISPR protein | | Descriptor: | Anti-CRISPR protein, AcrIE7 | | Authors: | Kang, J, Bae, E, Lee, G, Koo, J, Oh, H. | | Deposit date: | 2025-01-22 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Structural Investigation of the Anti-CRISPR Protein AcrIE7.
Proteins, 2025
|
|
9QH3
 
 | |
9QH0
 
 | |
4UHJ
 
 | |
6EK1
 
 | | Crystal structure of Type IIP restriction endonuclease PfoI | | Descriptor: | 1,2-ETHANEDIOL, restriction endonuclease PfoI | | Authors: | Tamulaitiene, G, Manakova, E, Jovaisaite, V, Grazulis, S, Siksnys, V. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Unique mechanism of target recognition by PfoI restriction endonuclease of the CCGG-family.
Nucleic Acids Res., 47, 2019
|
|
2VDJ
 
 | | Crystal Structure of Homoserine O-acetyltransferase (metA) from Bacillus Cereus with Homoserine | | Descriptor: | HOMOSERINE O-SUCCINYLTRANSFERASE, L-HOMOSERINE, SULFATE ION | | Authors: | Zubieta, C, Arkus, K.A.J, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2007-10-10 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Single Amino Acid Change is Responsible for Evolution of Acyltransferase Specificity in Bacterial Methionine Biosynthesis.
J.Biol.Chem., 283, 2008
|
|
4UHS
 
 | |
4UF0
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with epitherapuetic compound 2-(((2-((2-(dimethylamino)ethyl) (ethyl)amino)-2-oxoethyl)amino)methyl)isonicotinic acid. | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-{[(E)-2-(dimethylamino)ethenyl](ethyl)amino}-2-oxoethyl)amino]methyl}pyridine-4-carboxylic acid, FE (II) ION, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Tobias, K, Kopec, J, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2014-12-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
4UM4
 
 | |
4UHT
 
 | |
4UHK
 
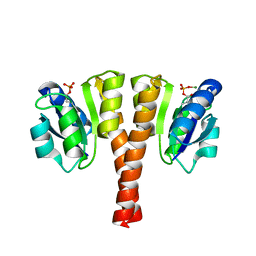 | |
6L9Z
 
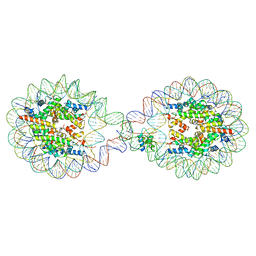 | | 338 bp di-nucleosome assembled with linker histone H1.X | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (338-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LAB
 
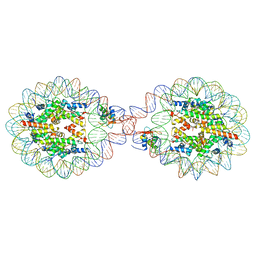 | | 169 bp nucleosome, harboring cohesive DNA termini, assembled with linker histone H1.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (169-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Bao, Q, Lee, P.L, Padavattan, S, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LA2
 
 | | 343 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | Descriptor: | DNA (343-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LER
 
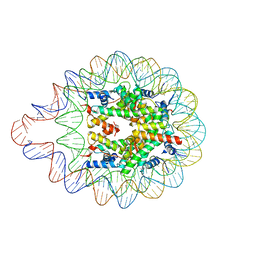 | | 169 bp nucleosome harboring non-identical cohesive DNA termini. | | Descriptor: | CALCIUM ION, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Sharma, D, Adhireksan, Z, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-26 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
4V48
 
 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the initiation-like state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
9MTR
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site GGS-mutant Release Factor 1, and P-site fMEAAAKC-peptidyl-tRNAcys at 2.80A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9MTQ
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site GGD-mutant Release Factor 1, and P-site fMEAAAKC-peptidyl-tRNAcys at 2.55A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9MTS
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site Q230-unmodified Release Factor 1, and P-site fMEAAAKC-peptidyl-tRNAcys at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9NO7
 
 | | Cryo-EM structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site Q230-N5-methylated Release Factor 1, and P-site 2'-deoxy-A76-fMEAAAKC-peptidyl-tRNAcys at 2.13A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-03-08 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9MTP
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site Q230-N5-methylated Release Factor 1, and P-site fMEAAAKC-peptidyl-tRNAcys at 2.40A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9MTT
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site Q230-N5-methylated Release Factor 1, and P-site deacylated-tRNAcys at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9ODR
 
 | | Structure of CRBN TBD bound to compound C1 | | Descriptor: | (3S)-3-(6-oxo-6,8-dihydro-2H,7H-spiro[furo[2,3-e]isoindole-3,4'-piperidin]-7-yl)piperidine-2,6-dione, Protein cereblon, ZINC ION | | Authors: | Strickland, C, Rice, C. | | Deposit date: | 2025-04-27 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery and Characterization of PVTX-321 as a Potent and Orally Bioavailable Estrogen Receptor Degrader for ER+/HER2- Breast Cancer.
J.Med.Chem., 68, 2025
|
|
4U30
 
 | |
6TD7
 
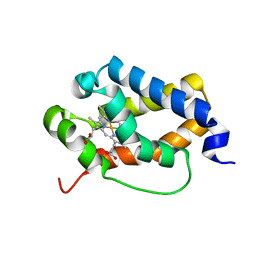 | | Structure of truncated hemoglobin THB11 from Chlamydomonas reinhardtii | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, THB11 | | Authors: | Huwald, D, Gasper, R, Hemschemeier, A, Hofmann, E. | | Deposit date: | 2019-11-08 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Distinctive structural properties of THB11, a pentacoordinate Chlamydomonas reinhardtii truncated hemoglobin with N- and C-terminal extensions.
J.Biol.Inorg.Chem., 25, 2020
|
|
