4Q9W
 
 | |
2HQK
 
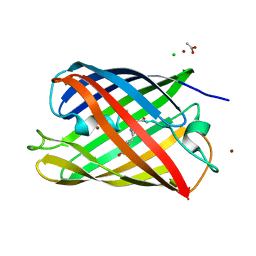 | | Crystal structure of a monomeric cyan fluorescent protein derived from Clavularia | | Descriptor: | ACETATE ION, CHLORIDE ION, Cyan fluorescent chromoprotein, ... | | Authors: | Henderson, J.N, Campbell, R.E, Ai, H, Remington, S.J. | | Deposit date: | 2006-07-18 | | Release date: | 2007-01-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging.
Biochem.J., 400, 2006
|
|
2OTE
 
 | | Crystal structure of a monomeric cyan fluorescent protein in the photobleached state | | Descriptor: | ACETATE ION, GFP-like fluorescent chromoprotein cFP484 | | Authors: | Henderson, J.N, Ai, H, Campbell, R.E, Remington, S.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for reversible photobleaching of a green fluorescent protein homologue.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4R6D
 
 | |
6FP7
 
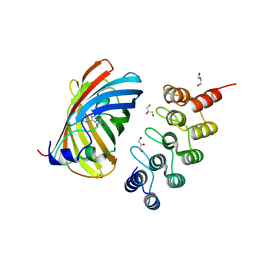 | | mTFP1/DARPin 1238_E11 complex in space group P6522 | | Descriptor: | DARPin1238_E11, GFP-like fluorescent chromoprotein cFP484, GLYCEROL | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6QSL
 
 | |
6QSM
 
 | |
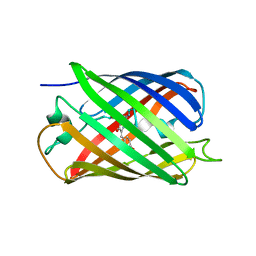
2OTB
 
 | |
6QSO
 
 | |
6FP8
 
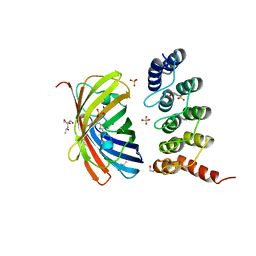 | | mTFP1/DARPin 1238_E11 complex in space group C2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DARPin1238_E11, ... | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
4Q9X
 
 | | mTFP* PdCl2 soak | | Descriptor: | CARBONATE ION, CHLORIDE ION, GFP-like fluorescent chromoprotein cFP484, ... | | Authors: | Fischer, J, Quitterer, F, Groll, M, Eppinger, J. | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | mTFP*: An imperishable and versatile protein host for anchoring diverse ligands and organocatalysts
To be Published
|
|
7WLD
 
 | | Cryo-EM structure of the human glycosylphosphatidylinositol transamidase complex at 2.53 Angstrom resolution | | Descriptor: | (4S,7R)-7-[(hexadecanoyloxy)methyl]-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-3,5,8-trioxa-4lambda~5~-phosphahexacosan-1-aminium, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, Y, Li, T, Luo, Y, Chao, Y, Jia, G, Zhou, Z, Su, Z, Qu, Q, Li, D. | | Deposit date: | 2022-01-13 | | Release date: | 2022-04-27 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Molecular insights into biogenesis of glycosylphosphatidylinositol anchor proteins.
Nat Commun, 13, 2022
|
|
8IMX
 
 | | Cryo-EM structure of GPI-T with a chimeric GPI-anchored protein | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, Y, Li, T, Qu, Q, Li, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
7W72
 
 | | Structure of a human glycosylphosphatidylinositol (GPI) transamidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GPI transamidase component PIG-S, ... | | Authors: | Zhang, H, Su, J, Li, B, Gao, Y, Zhang, X.C, Zhao, Y. | | Deposit date: | 2021-12-02 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of human glycosylphosphatidylinositol transamidase.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8IMY
 
 | | Cryo-EM structure of GPI-T (inactive mutant) with GPI and proULBP2, a proprotein substrate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, T, Xu, Y, Qu, Q, Li, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
