8A5I
 
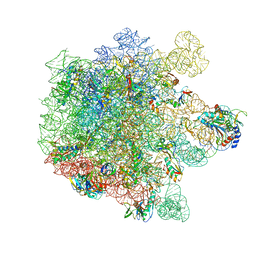 | | Cryo-EM structure of Lincomycin bound to the Listeria monocytogenes 50S ribosomal subunit. | | Descriptor: | 1,4-DIAMINOBUTANE, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Koller, T.O, Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2022-06-15 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for HflXr-mediated antibiotic resistance in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
8A57
 
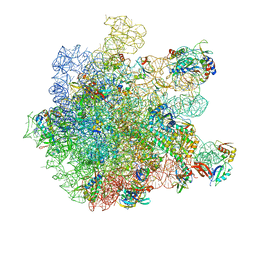 | | Cryo-EM structure of HflXr bound to the Listeria monocytogenes 50S ribosomal subunit. | | Descriptor: | 1,4-DIAMINOBUTANE, 23S ribosomal RNA, 50S ribosomal protein L11, ... | | Authors: | Koller, T.O, Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2022-06-14 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for HflXr-mediated antibiotic resistance in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
7NHN
 
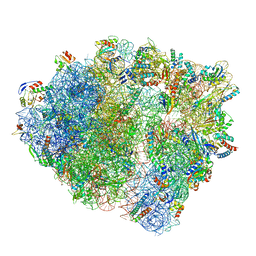 | | VgaL, an antibiotic resistance ABCF, in complex with 70S ribosome from Listeria monocytogenes | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Turnbull, K.J, Hauryliuk, V, Wilson, D.N. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens.
Nat Commun, 12, 2021
|
|
8A63
 
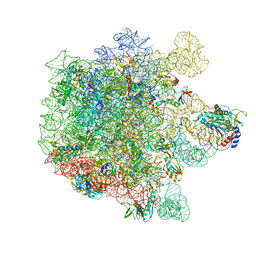 | | Cryo-EM structure of Listeria monocytogenes 50S ribosomal subunit. | | Descriptor: | 1,4-DIAMINOBUTANE, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Koller, T.O, Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for HflXr-mediated antibiotic resistance in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
