2XI8
 
 | | High resolution structure of native CylR2 | | Descriptor: | GLYCEROL, PUTATIVE TRANSCRIPTION REGULATOR | | Authors: | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-06-28 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
2XJ3
 
 | | High resolution structure of the T55C mutant of CylR2. | | Descriptor: | CYLR2 SYNONYM CYTOLYSIN REPRESSOR 2, GLYCEROL | | Authors: | Gruene, T, Cho, M.K, Karyagina, I, Kim, H.Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-02 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
2XIU
 
 | | High resolution structure of MTSL-tagged CylR2. | | Descriptor: | CYLR2, GLYCEROL, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
1UTX
 
 | | Regulation of Cytolysin Expression by Enterococcus faecalis: Role of CylR2 | | Descriptor: | CYLR2, IODIDE ION, SODIUM ION | | Authors: | Razeto, A, Rumpel, S, Pillar, C.M, Gilmore, M.S, Becker, S, Zweckstetter, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-09-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and DNA-Binding Properties of the Cytolysin Regulator CylR2 from Enterococcus Faecalis
Embo J., 23, 2004
|
|
2LYP
 
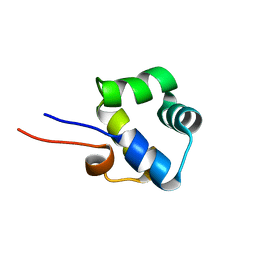 | | NOE-based 3D structure of the monomer of CylR2 in equilibrium with predissociated homodimer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYJ
 
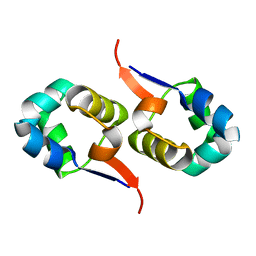 | | NOE-based 3D structure of the CylR2 homodimer at 298K | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Giller, K, Becker, S, Zweckstetter, M, Schwieters, C.D. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYK
 
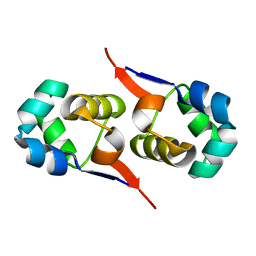 | | NOE-based 3D structure of the CylR2 homodimer at 270K (-3 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYQ
 
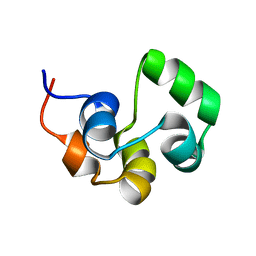 | | NOE-based 3D structure of the monomeric intermediate of CylR2 at 262K (-11 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYL
 
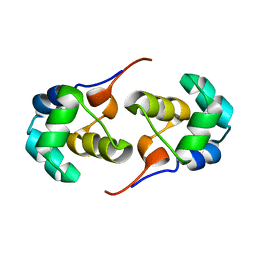 | | NOE-based 3D structure of the predissociated homodimer of CylR2 in equilibrium with monomer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYS
 
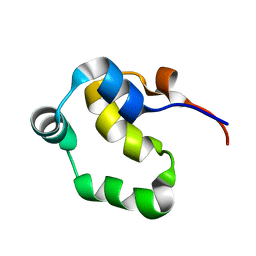 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 257K (-16 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYR
 
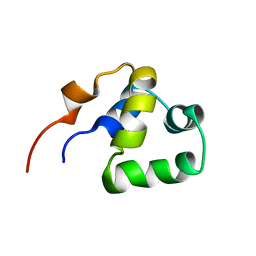 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 259K (-14 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2GZU
 
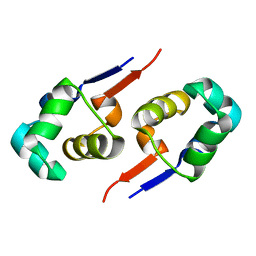 | |
