2QVE
 
 | |
2RJR
 
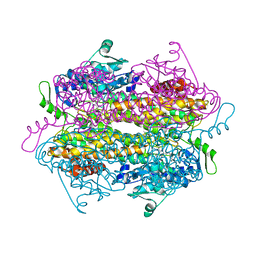 | | Substrate mimic bound to SgTAM | | Descriptor: | (2S,3S)-3-(4-fluorophenyl)-2,3-dihydroxypropanoic acid, Tyrosine aminomutase | | Authors: | Montavon, T.J, Christianson, C.V, Bruner, S.D. | | Deposit date: | 2007-10-15 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and characterization of mechanism-based inhibitors for the tyrosine aminomutase SgTAM.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3KDY
 
 | |
3KDZ
 
 | |
2RJS
 
 | | SgTAM bound to substrate mimic | | Descriptor: | (3R)-3-amino-2,2-difluoro-3-(4-methoxyphenyl)propanoic acid, Tyrosine aminomutase | | Authors: | Montavon, T.J, Christianson, C.V, Bruner, S.D. | | Deposit date: | 2007-10-15 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and characterization of mechanism-based inhibitors for the tyrosine aminomutase SgTAM.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2OHY
 
 | |
