1F1S
 
 | |
1I8Q
 
 | |
1LXM
 
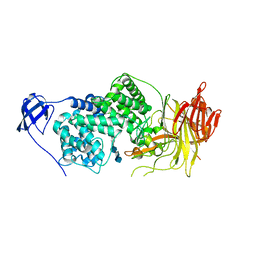 | | Crystal Structure of Streptococcus agalactiae Hyaluronate Lyase Complexed with Hexasaccharide Unit of Hyaluronan | | Descriptor: | HYALURONATE Lyase, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mello, L.V, de Groot, B.L, Li, S, Jedrzejas, M.J. | | Deposit date: | 2002-06-05 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and flexibility of Streptococcus agalactiae hyaluronate lyase complex with its substrate. Insights into the mechanism of processive degradation of hyaluronan.
J.Biol.Chem., 277, 2002
|
|
