9Q8F
 
 | | Structure of the (6-4) photolyase of Caulobacter crescentus with K48A mutation in its dark adapted and oxidized state determined by synchrotron | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, CHLORIDE ION, Cryptochrome/photolyase family protein, ... | | Authors: | Po Hsun, W, Maestre-Reyna, M, Essen, L.-O. | | Deposit date: | 2025-02-24 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-State-Dependent Structural Changes within a Prokaryotic 6-4 Photolyase.
J.Am.Chem.Soc., 147, 2025
|
|
8IJY
 
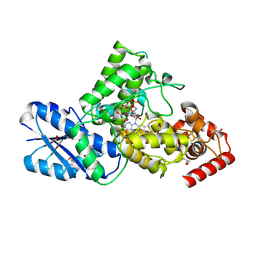 | | Synechococcus elongatus 6-4 photolyase with an 8-HDF as the antenna chromophore and a covalently linked FAD as the catalytic cofactor | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, Deoxyribodipyrimidine photolyase-related protein, ... | | Authors: | Liu, Y, Xu, L, Zhang, P. | | Deposit date: | 2023-02-28 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Synechococcus elongatus 6-4 photolyase with an 8-HDF as the antenna chromophore and a covalently linked FAD as the catalytic cofactor
To Be Published
|
|
6DD6
 
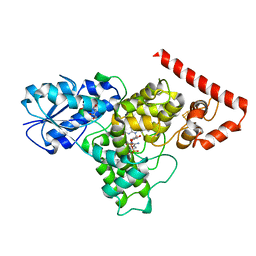 | |
3ZXS
 
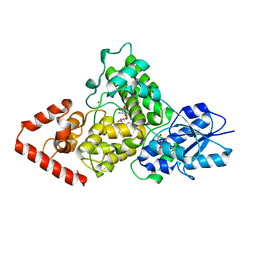 | | Cryptochrome B from Rhodobacter sphaeroides | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, CRYPTOCHROME B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Geisselbrecht, Y, Fruhwirth, S, Pierik, A.J, Klug, G, Essen, L.-O. | | Deposit date: | 2011-08-15 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cryb from Rhodobacter Sphaeroides: A Unique Class of Cryptochromes with New Cofactors.
Embo Rep., 13, 2012
|
|
4DJA
 
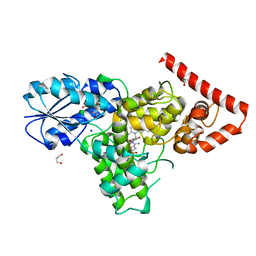 | | Crystal structure of a prokaryotic (6-4) photolyase PhrB from Agrobacterium Tumefaciens with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore at 1.45A resolution | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Scheerer, P, Zhang, F, Oberpichler, I, Lamparter, T, Krauss, N. | | Deposit date: | 2012-02-01 | | Release date: | 2013-04-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a prokaryotic (6-4) photolyase with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5KCM
 
 | | Crystal structure of iron-sulfur cluster containing photolyase PhrB mutant I51W | | Descriptor: | (6-4) photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yang, X, Bowatte, K, Zhang, F, Lamparter, T. | | Deposit date: | 2016-06-06 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
5LFA
 
 | | Crystal structure of iron-sulfur cluster containing bacterial (6-4) photolyase PhrB - Y424F mutant with impaired DNA repair activity | | Descriptor: | (6-4) photolyase, 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kwiatkowski, D, Zhang, F, Krauss, N, Lamparter, T, Scheerer, P. | | Deposit date: | 2016-06-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
8A1H
 
 | | Bacterial 6-4 photolyase from Vibrio cholerase | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-4 photolyase (FeS-BCP, ... | | Authors: | Essen, L.-O, Emmerich, H.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Functional Analysis of a Prokaryotic (6-4) Photolyase from the Aquatic Pathogen Vibrio Cholerae † .
Photochem.Photobiol., 99, 2023
|
|
7YKN
 
 | | Crystal structure of (6-4) photolyase from Vibrio cholerae | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Cryptochrome/photolyase family protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cakilkaya, B, Kavakli, I.H, DeMirci, H. | | Deposit date: | 2022-07-23 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of Vibrio cholerae (6-4) photolyase reveals interactions with cofactors and a DNA-binding region.
J.Biol.Chem., 299, 2023
|
|
9HNL
 
 | | Structure of the (6-4) photolyase of Caulobacter crescentus in its oxidized state at room temperture-synchrotron | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Cryptochrome/photolyase family protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Po Hsun, W, Maestre-Reyna, M, Essen, L.-O. | | Deposit date: | 2024-12-11 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Redox-State-Dependent Structural Changes within a Prokaryotic 6-4 Photolyase.
J.Am.Chem.Soc., 147, 2025
|
|
9HNN
 
 | | Structure of the (6-4) photolyase of Caulobacter crescentus in its fully reduced state determined by serial femtosecond crystallography | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Cryptochrome/photolyase family protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Po Hsun, W, Maestre-Reyna, M, Essen, L.-O. | | Deposit date: | 2024-12-11 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Redox-State-Dependent Structural Changes within a Prokaryotic 6-4 Photolyase.
J.Am.Chem.Soc., 147, 2025
|
|
9HNK
 
 | | Structure of the (6-4) photolyase of Caulobacter crescentus in its oxidizd state - Singal crystal / Synchrotron | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Cryptochrome/photolyase family protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Po Hsun, W, Maestre-Reyna, M, Essen, L.-O. | | Deposit date: | 2024-12-11 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Redox-State-Dependent Structural Changes within a Prokaryotic 6-4 Photolyase.
J.Am.Chem.Soc., 147, 2025
|
|
9HNM
 
 | | Structure of the (6-4) photolyase of Caulobacter crescentus in its dark adapted and oxidized state determined by serial femtosecond crystallography | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Cryptochrome/photolyase family protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Po Hsun, W, Maestre-Reyna, M, Essen, L.-O. | | Deposit date: | 2024-12-11 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Redox-State-Dependent Structural Changes within a Prokaryotic 6-4 Photolyase.
J.Am.Chem.Soc., 147, 2025
|
|
9HNO
 
 | | Structure of the (6-4) photolyase of Caulobacter crescentus in its iron sulfur cluster oxidized state determined by serial femtosecond crystallography | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Cryptochrome/photolyase family protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Po Hsun, W, Maestre-Reyna, M, Essen, L.-O. | | Deposit date: | 2024-12-11 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redox-State-Dependent Structural Changes within a Prokaryotic 6-4 Photolyase.
J.Am.Chem.Soc., 147, 2025
|
|
