5U8E
 
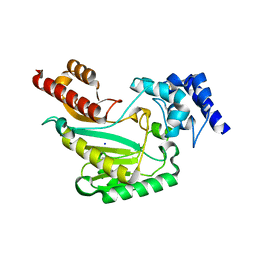 | | Crystal Structure of substrate-free arginine kinase from spider Polybetes pythagoricus | | Descriptor: | SODIUM ION, arginine kinase | | Authors: | Lopez-Zavala, A.A, Garcia, C.F, Hernadez-Paredes, J, Sotelo-Mundo, R.R. | | Deposit date: | 2016-12-14 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Biochemical and structural characterization of a novel arginine kinase from the spider Polybetes pythagoricus.
PeerJ, 5, 2017
|
|
1I0E
 
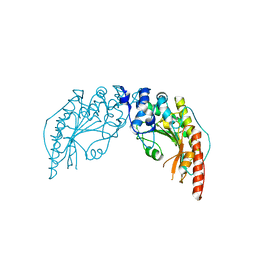 | | CRYSTAL STRUCTURE OF CREATINE KINASE FROM HUMAN MUSCLE | | Descriptor: | CREATINE KINASE,M CHAIN | | Authors: | Shen, Y.-Q, Tang, L, Zhou, H.-M, Lin, Z.-J. | | Deposit date: | 2001-01-29 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of human muscle creatine kinase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
8CI4
 
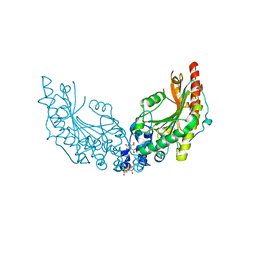 | |
3JU6
 
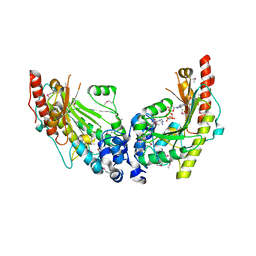 | | Crystal Structure of Dimeric Arginine Kinase in Complex with AMPPNP and Arginine | | Descriptor: | ARGININE, Arginine kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wu, X, Ye, S, Guo, S, Yan, W, Bartlam, M, Rao, Z. | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for a reciprocating mechanism of negative cooperativity in dimeric phosphagen kinase activity
Faseb J., 24, 2010
|
|
1U6R
 
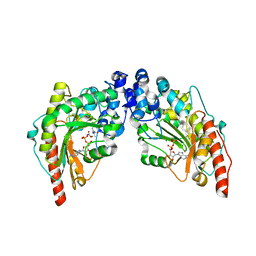 | | Transition state analog complex of muscle creatine kinase (R134K) mutant | | Descriptor: | (DIAMINOMETHYL-METHYL-AMINO)-ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, Creatine kinase, ... | | Authors: | Ohren, J.F, Kundracik, M.L, Borders, C.L, Edmiston, P, Viola, R.E. | | Deposit date: | 2004-07-30 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural asymmetry and intersubunit communication in muscle creatine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
7EWS
 
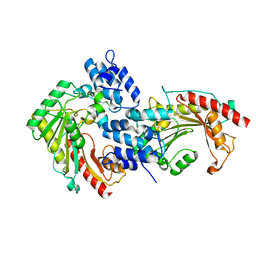 | | Crystal structure of arginine kinase (AK3) from the ciliate Paramecium tetraurelia | | Descriptor: | arginine kinase | | Authors: | Otsuka, Y, Yokota, J, Yano, D, Uda, K, Suzuki, T, Sugiyama, S. | | Deposit date: | 2021-05-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of arginine kinase (AK3) from the ciliate Paramecium tetraurelia
To Be Published
|
|
4V7N
 
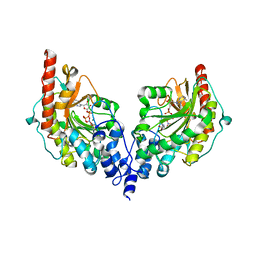 | | Glycocyamine kinase, beta-beta homodimer from marine worm Namalycastis sp., with transition state analog Mg(II)-ADP-NO3-glycocyamine. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANIDINO ACETATE, Glycocyamine kinase beta chain, ... | | Authors: | Lim, K, Pullalarevu, S, Herzberg, O. | | Deposit date: | 2009-12-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the mechanism and substrate specificity of glycocyamine kinase, a phosphagen kinase family member.
Biochemistry, 49, 2010
|
|
6V9H
 
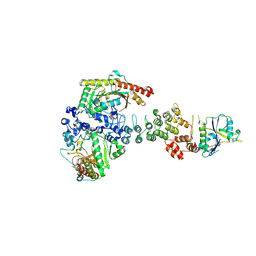 | | Ankyrin repeat and SOCS-box protein 9 (ASB9), ElonginB (ELOB), and ElonginC (ELOC) bound to its substrate Brain-type Creatine Kinase (CKB) | | Descriptor: | Ankyrin repeat and SOCS box protein 9, Creatine kinase B-type, Elongin-B, ... | | Authors: | Komives, E.A, Lumpkin, R.J, Baker, R.W, Leschziner, A.E. | | Deposit date: | 2019-12-13 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and dynamics of the ASB9 CUL-RING E3 Ligase.
Nat Commun, 11, 2020
|
|
4Z9M
 
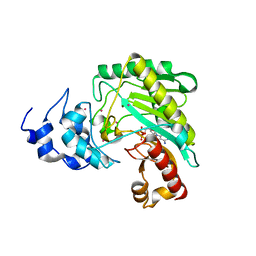 | | Crystal structure of human sarcomeric mitochondrial creatine kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase S-type, mitochondrial, ... | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-04-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human sarcomeric mitochondrial creatine kinase
To Be Published
|
|
7RE6
 
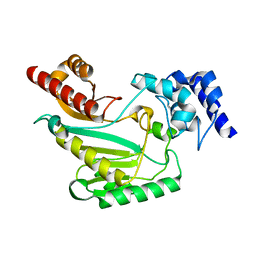 | |
1VRP
 
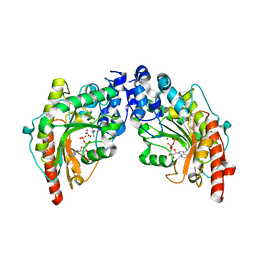 | | The 2.1 Structure of T. californica Creatine Kinase Complexed with the Transition-State Analogue Complex, ADP-Mg 2+ /NO3-/Creatine | | Descriptor: | (DIAMINOMETHYL-METHYL-AMINO)-ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, Creatine Kinase, ... | | Authors: | Lahiri, S.D, Wang, P.F, Babbitt, P.C, McLeish, M.J, Kenyon, G.L, Allen, K.N. | | Deposit date: | 2005-04-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 A Structure of Torpedo californica Creatine Kinase Complexed with the ADP-Mg(2+)-NO3(-)-Creatine Transition-State Analogue Complex
Biochemistry, 41, 2002
|
|
2J1Q
 
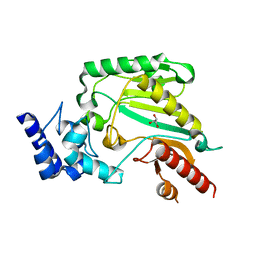 | | Crystal structure of Trypanosoma cruzi arginine kinase | | Descriptor: | ARGININE KINASE, GLYCEROL | | Authors: | Fernandez, P, Haouz, A, Pereira, C.A, Aguilar, C, Alzari, P.M. | | Deposit date: | 2006-08-15 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Trypanosoma Cruzi Arginine Kinase.
Proteins, 69, 2007
|
|
7TUN
 
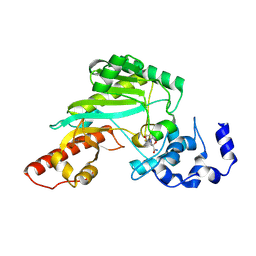 | |
3B6R
 
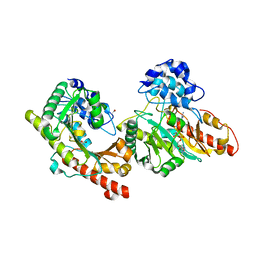 | | Crystal structure of Human Brain-type Creatine Kinase | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, Creatine kinase B-type, ... | | Authors: | Bong, S.M, Moon, J.H, Hwang, K.Y. | | Deposit date: | 2007-10-29 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of human brain-type creatine kinase complexed with the ADP-Mg2+-NO3- -creatine transition-state analogue complex
Febs Lett., 582, 2008
|
|
1BG0
 
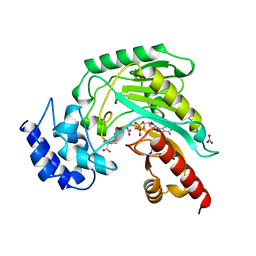 | | TRANSITION STATE STRUCTURE OF ARGININE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE KINASE, D-ARGININE, ... | | Authors: | Zhou, G, Somasundaram, T, Blanc, E, Parthasarathy, G, Ellington, W.R, Chapman, M.S. | | Deposit date: | 1998-06-03 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Transition state structure of arginine kinase: implications for catalysis of bimolecular reactions.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
5ZHQ
 
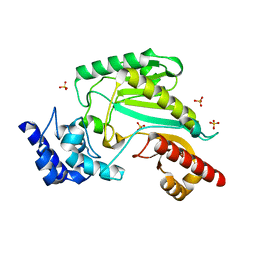 | |
5U92
 
 | | Crystal Structure of arginine kinase from the spider Polybetes pythagoricus in complex with arginine | | Descriptor: | ARGININE, SODIUM ION, arginine kinase | | Authors: | Lopez-zavala, A.A, Garcia, C.F, Paredes-Hernandez, J, Stojanoff, V, Sotelo-Mundo, R.R. | | Deposit date: | 2016-12-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of a novel arginine kinase from the spider Polybetes pythagoricus.
PeerJ, 5, 2017
|
|
2CRK
 
 | | MUSCLE CREATINE KINASE | | Descriptor: | PROTEIN (CREATINE KINASE), SULFATE ION | | Authors: | Rao, J.K, Bujacz, G, Wlodawer, A. | | Deposit date: | 1998-09-28 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of rabbit muscle creatine kinase.
FEBS Lett., 439, 1998
|
|
7VCJ
 
 | | Arginine kinase H227A from Daphnia magna | | Descriptor: | Arginine kinase, NITRATE ION, PHOSPHATE ION | | Authors: | Kim, D.S, Jang, K, Kim, W.S, Kim, Y.J, Park, J.H. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of H227A Mutant of Arginine Kinase in Daphnia magna Suggests the Importance of Its Stability.
Molecules, 27, 2022
|
|
3DRE
 
 | |
3DRB
 
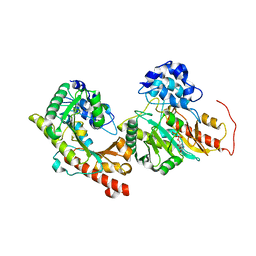 | | Crystal structure of Human Brain-type Creatine Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase B-type, MAGNESIUM ION | | Authors: | Moon, J.H, Bong, S.M, Hwang, K.Y, Chi, Y.M. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of human brain-type creatine kinase complexed with the ADP-Mg2+-NO3- -creatine transition-state analogue complex
Febs Lett., 582, 2008
|
|
7U5I
 
 | |
5J9A
 
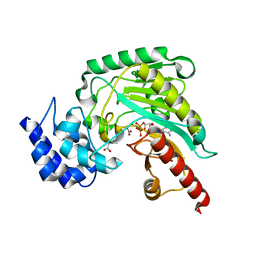 | | Ambient temperature transition state structure of arginine kinase - crystal 11/Form II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Godsey, M, Davulcu, O, Nix, J, Skalicky, J.J, Bruschweiler, R, Chapman, M.S. | | Deposit date: | 2016-04-08 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | The Sampling of Conformational Dynamics in Ambient-Temperature Crystal Structures of Arginine Kinase.
Structure, 24, 2016
|
|
1CRK
 
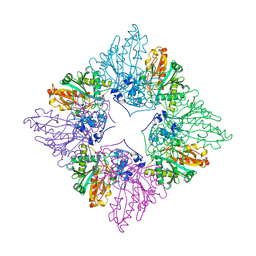 | | MITOCHONDRIAL CREATINE KINASE | | Descriptor: | CREATINE KINASE, PHOSPHATE ION | | Authors: | Fritz-Wolf, K, Schnyder, T, Wallimann, T, Kabsch, W. | | Deposit date: | 1996-03-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of mitochondrial creatine kinase.
Nature, 381, 1996
|
|
5J99
 
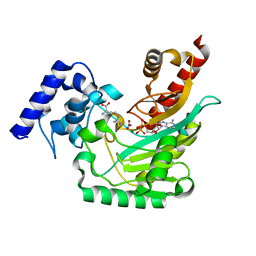 | | Ambient temperature transition state structure of arginine kinase - crystal 8/Form I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Godsey, M, Davulcu, O, Nix, J, Skalicky, J.J, Bruschweiler, R, Chapman, M.S. | | Deposit date: | 2016-04-08 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Sampling of Conformational Dynamics in Ambient-Temperature Crystal Structures of Arginine Kinase.
Structure, 24, 2016
|
|
