6K37
 
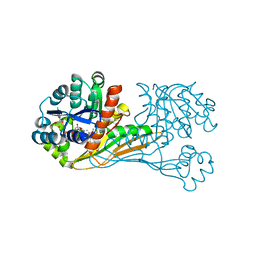 | | Crystal structure of BioU (K124A) from Synechocystis sp.PCC6803 in complex with NAD+ and the analog of reaction intermediate, 3-(1-aminoethyl)-nonanedioic acid | | Descriptor: | (3R)-3-[(1R)-1-azanylethyl]nonanedioic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Slr0355 protein | | Authors: | Sakaki, K, Tomita, T, Nishiyama, M. | | Deposit date: | 2019-05-16 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria.
Nat.Chem.Biol., 16, 2020
|
|
6K38
 
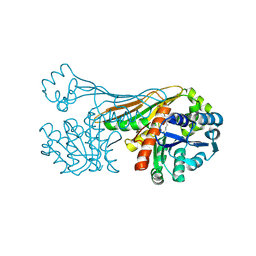 | |
6ITD
 
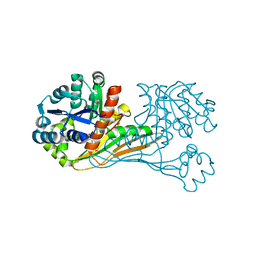 | | Crystal structure of BioU (K124A) from Synechocystis sp.PCC6803 in complex with the analog of reaction intermediate, 3-(1-aminoethyl)-nonanedioic acid | | Descriptor: | 3-(1-AMINOETHYL)NONANEDIOIC ACID, Slr0355 protein | | Authors: | Sakaki, K, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2018-11-21 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria.
Nat.Chem.Biol., 16, 2020
|
|
6IR4
 
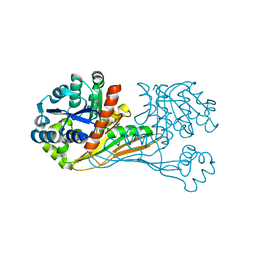 | | Crystal structure of BioU from Synechocystis sp.PCC6803 (apo form) | | Descriptor: | Slr0355 protein | | Authors: | Sakaki, K, Oishi, K, Shimizu, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2018-11-10 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria.
Nat.Chem.Biol., 16, 2020
|
|
6K36
 
 | |
5B37
 
 | | Crystal structure of L-tryptophan dehydrogenase from Nostoc punctiforme | | Descriptor: | Tryptophan dehydrogenase | | Authors: | Wakamatsu, T, Sakuraba, H, Kitamura, M, Hakumai, Y, Ohnishi, K, Ashiuchi, M, Ohshima, T. | | Deposit date: | 2016-02-11 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Insights into l-Tryptophan Dehydrogenase from a Photoautotrophic Cyanobacterium, Nostoc punctiforme.
Appl. Environ. Microbiol., 83, 2017
|
|
4XGI
 
 | |
3JD2
 
 | | Glutamate dehydrogenase in complex with NADH, open conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
2TMG
 
 | | THERMOTOGA MARITIMA GLUTAMATE DEHYDROGENASE MUTANT S128R, T158E, N117R, S160E | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Knapp, S, Lebbink, J.H.G, van der Oost, J, de Vos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering activity and stability of Thermotoga maritima glutamate dehydrogenase. II: construction of a 16-residue ion-pair network at the subunit interface.
J.Mol.Biol., 289, 1999
|
|
3VPX
 
 | | Crystal structure of leucine dehydrogenase from a psychrophilic bacterium Sporosarcina psychrophila. | | Descriptor: | Leucine dehydrogenase | | Authors: | Zhao, Y, Wakamatsu, T, Doi, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2012-03-14 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A psychrophilic leucine dehydrogenase from Sporosarcina psychrophila: Purification, characterization, gene sequencing and crystal structure analysis
J.MOL.CATAL., B ENZYM., 83, 2012
|
|
2BMA
 
 | | The crystal structure of Plasmodium falciparum glutamate dehydrogenase, a putative target for novel antimalarial drugs | | Descriptor: | GLUTAMATE DEHYDROGENASE (NADP+) | | Authors: | Werner, C, Stubbs, M.T, Krauth-Siege, R.L, Klebe, G. | | Deposit date: | 2005-03-10 | | Release date: | 2005-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Plasmodium Falciparum Glutamate Dehydrogenase, a Putative Target for Novel Antimalarial Drugs
J.Mol.Biol., 349, 2005
|
|
8EW0
 
 | |
5K12
 
 | | Cryo-EM structure of glutamate dehydrogenase at 1.8 A resolution | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.8 Å) | | Cite: | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
5IJZ
 
 | |
4BHT
 
 | | Structural Determinants of Cofactor Specificity and Domain Flexibility in Bacterial Glutamate Dehydrogenases | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, NADP-SPECIFIC GLUTAMATE DEHYDROGENASE, ... | | Authors: | Oliveira, T, Sharkey, M, Engel, P, Khan, A. | | Deposit date: | 2013-04-06 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Nadp(+) -Dependent Glutamate Dehydrogenase from Escherichia Coli: Reflections on the Basis of Coenzyme Specificity in the Family of Glutamate Dehydrogenases.
FEBS J., 280, 2013
|
|
1V9L
 
 | | L-glutamate dehydrogenase from Pyrobaculum islandicum complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, glutamate dehydrogenase | | Authors: | Bhuiya, M.W, Sakuraba, H, Ohshima, T, Imagawa, T, Katunuma, N, Tsuge, H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The First Crystal Structure of Hyperthermostable NAD-dependent Glutamate Dehydrogenase from Pyrobaculum islandicum
J.Mol.Biol., 345, 2005
|
|
8AR7
 
 | |
8AR8
 
 | |
6YEH
 
 | | Arabidopsis thaliana glutamate dehydrogenase isoform 1 in apo form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glutamate dehydrogenase 1, POTASSIUM ION | | Authors: | Ruszkowski, M, Grzechowiak, M, Jaskolski, M. | | Deposit date: | 2020-03-24 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Studies of Glutamate Dehydrogenase (Isoform 1) FromArabidopsis thaliana, an Important Enzyme at the Branch-Point Between Carbon and Nitrogen Metabolism.
Front Plant Sci, 11, 2020
|
|
6YEI
 
 | | Arabidopsis thaliana glutamate dehydrogenase isoform 1 in complex with NAD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Ruszkowski, M, Grzechowiak, M, Jaskolski, M. | | Deposit date: | 2020-03-24 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Studies of Glutamate Dehydrogenase (Isoform 1) FromArabidopsis thaliana, an Important Enzyme at the Branch-Point Between Carbon and Nitrogen Metabolism.
Front Plant Sci, 11, 2020
|
|
4FCC
 
 | | Glutamate dehydrogenase from E. coli | | Descriptor: | Glutamate dehydrogenase | | Authors: | Bilokapic, S, Schwartz, T.U. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7MFT
 
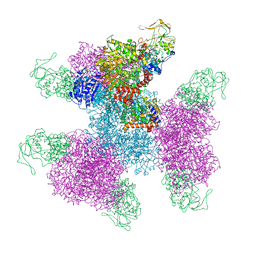 | | Glutamate synthase, glutamate dehydrogenase counter-enzyme complex (GudB6-GltA6-GltB6) | | Descriptor: | FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Jayaraman, V, Lee, D.J, Elad, N, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2021-04-11 | | Release date: | 2022-01-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A counter-enzyme complex regulates glutamate metabolism in Bacillus subtilis.
Nat.Chem.Biol., 18, 2022
|
|
7MFM
 
 | | Glutamate synthase, glutamate dehydrogenase counter-enzyme complex | | Descriptor: | FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Jayaraman, V, Lee, D.J, Elad, N, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2021-04-10 | | Release date: | 2022-01-05 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | A counter-enzyme complex regulates glutamate metabolism in Bacillus subtilis.
Nat.Chem.Biol., 18, 2022
|
|
4FHN
 
 | |
3SBO
 
 | | Structure of E.coli GDH from native source | | Descriptor: | CHLORIDE ION, NADP-specific glutamate dehydrogenase | | Authors: | Gee, C.L, Zubieta, C, Echols, N, Totir, M. | | Deposit date: | 2011-06-06 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
