1LM4
 
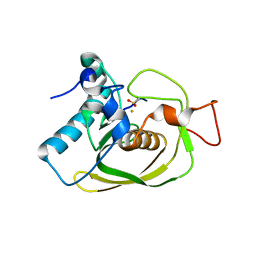 | | Structure of Peptide Deformylase from Staphylococcus aureus at 1.45 A | | Descriptor: | FE (III) ION, GLYCEROL, peptide deformylase PDF1 | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-04-30 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
6JFO
 
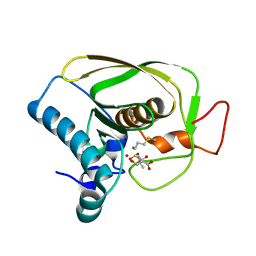 | |
1LQW
 
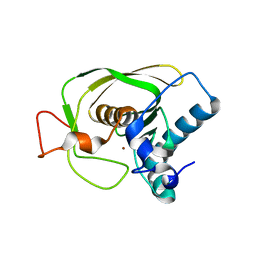 | | Crystal Structure of S.aureus Peptide Deformylase | | Descriptor: | PEPTIDE DEFORMYLASE PDF1, ZINC ION | | Authors: | Mikol, V. | | Deposit date: | 2002-05-14 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents.
J.Mol.Biol., 320, 2002
|
|
1LMH
 
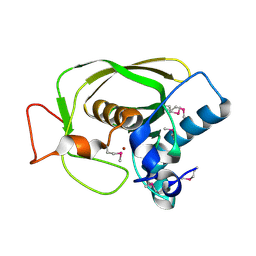 | |
1Q1Y
 
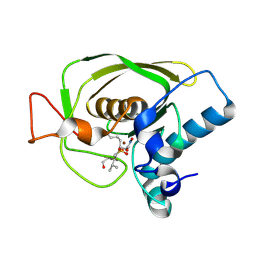 | | Crystal Structures of Peptide Deformylase from Staphylococcus aureus Complexed with Actinonin | | Descriptor: | ACTINONIN, Peptide deformylase, ZINC ION | | Authors: | Yoon, H.J, Lee, S.K, Kim, H.L, Kim, H.W, Kim, H.W, Lee, J.Y, Mikami, B, Suh, S.W. | | Deposit date: | 2003-07-23 | | Release date: | 2004-07-23 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Proteins, 57, 2004
|
|
6JFQ
 
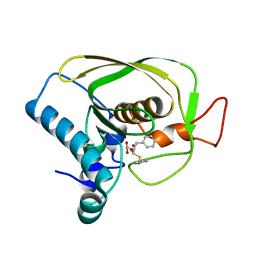 | |
6JFS
 
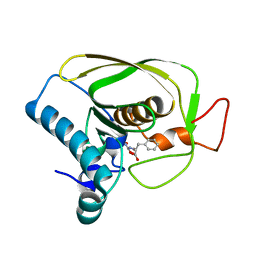 | |
6JFR
 
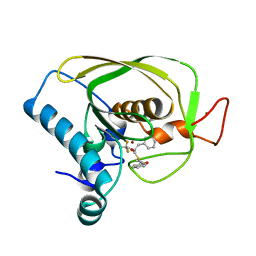 | |
2AI9
 
 | | S.aureus Polypeptide Deformylase | | Descriptor: | NICKEL (II) ION, Peptide deformylase, SULFATE ION | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species.
Protein Sci., 12, 2003
|
|
6JFG
 
 | |
