4YZG
 
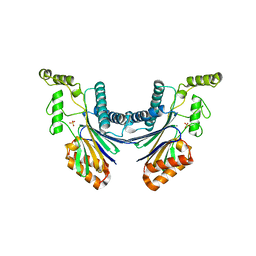 | | Structure of the Arabidopsis TAP38/PPH1, a state-transition phosphatase responsible for dephosphorylation of LHCII | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 2C 57, SULFATE ION | | Authors: | Wei, X.P, Guo, J.T, Li, M, Liu, Z.F. | | Deposit date: | 2015-03-25 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Mechanism Underlying the Specific Recognition between the Arabidopsis State-Transition Phosphatase TAP38/PPH1 and Phosphorylated Light-Harvesting Complex Protein Lhcb1
Plant Cell, 27, 2015
|
|
4YZH
 
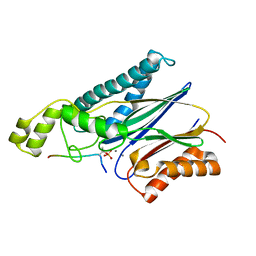 | | Structure of the Arabidopsis TAP38/PPH1 in complex with pLhcb1 phosphopeptide substrate | | Descriptor: | Chlorophyll a-b binding protein 2, chloroplastic, MANGANESE (II) ION, ... | | Authors: | Wei, X.P, Guo, J.T, Li, M, Liu, Z.F. | | Deposit date: | 2015-03-25 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Mechanism Underlying the Specific Recognition between the Arabidopsis State-Transition Phosphatase TAP38/PPH1 and Phosphorylated Light-Harvesting Complex Protein Lhcb1
Plant Cell, 27, 2015
|
|
