3BMX
 
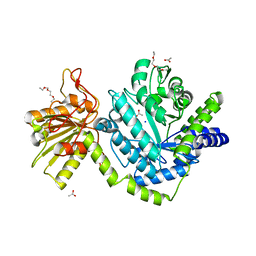 | | Beta-N-hexosaminidase (YbbD) from Bacillus subtilis | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ACETATE ION, SODIUM ION, ... | | Authors: | Fischer, S. | | Deposit date: | 2007-12-13 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and kinetic analysis of Bacillus subtilis N-acetylglucosaminidase reveals a unique Asp-His dyad mechanism
J.Biol.Chem., 285, 2010
|
|
4GYJ
 
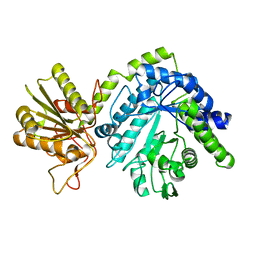 | |
4GYK
 
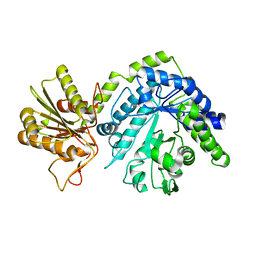 | |
3NVD
 
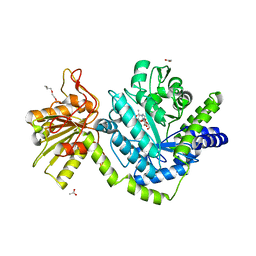 | | Structure of YBBD in complex with pugnac | | Descriptor: | ACETATE ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, SODIUM ION, ... | | Authors: | Diederichs, K. | | Deposit date: | 2010-07-08 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.836 Å) | | Cite: | Structural and kinetic analysis of Bacillus subtilis N-acetylglucosaminidase reveals a unique Asp-His dyad mechanism
J.Biol.Chem., 285, 2010
|
|
3LK6
 
 | |
