4IC4
 
 | |
4INQ
 
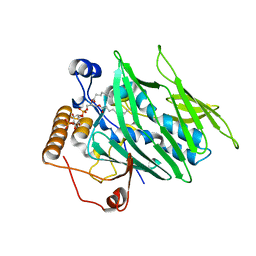 | | Crystal structure of Osh3 ORD in complex with PI(4)P from Saccharomyces cerevisiae | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, Oxysterol-binding protein homolog 3 | | Authors: | Tong, J, Im, Y.J. | | Deposit date: | 2013-01-05 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins
Structure, 21, 2013
|
|
4IAP
 
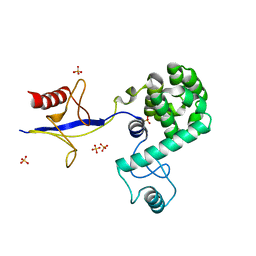 | | Crystal structure of PH domain of Osh3 from Saccharomyces cerevisiae | | Descriptor: | Oxysterol-binding protein homolog 3,Endolysin,Oxysterol-binding protein homolog 3, SULFATE ION | | Authors: | Tong, J, Im, Y.J. | | Deposit date: | 2012-12-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins
Structure, 21, 2013
|
|
