4I5U
 
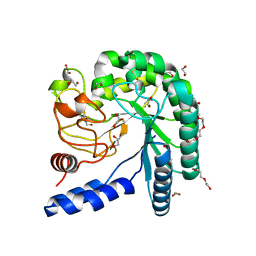 | | Crystal structure of a fungal chimeric cellobiohydrolase Cel6A | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Chimeric cel6A, ... | | Authors: | Arnold, F.H, Wu, I. | | Deposit date: | 2012-11-29 | | Release date: | 2013-04-03 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Engineered thermostable fungal Cel6A and Cel7A cellobiohydrolases hydrolyze cellulose efficiently at elevated temperatures.
Biotechnol.Bioeng., 110, 2013
|
|
4I5R
 
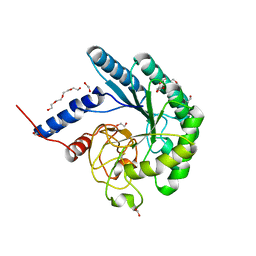 | | Crystal structure of a fungal chimeric cellobiohydrolase Cel6A | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Chimeric cel6A, ... | | Authors: | Arnold, F.H, Wu, I. | | Deposit date: | 2012-11-28 | | Release date: | 2013-04-03 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineered thermostable fungal Cel6A and Cel7A cellobiohydrolases hydrolyze cellulose efficiently at elevated temperatures.
Biotechnol.Bioeng., 110, 2013
|
|
4AX7
 
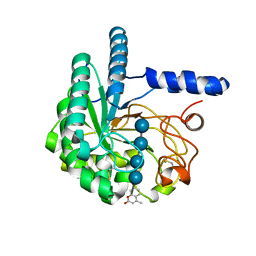 | | Hypocrea jecorina Cel6A D221A mutant soaked with 4-Methylumbelliferyl- beta-D-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-hydroxy-4-methyl-2H-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-06-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
4AU0
 
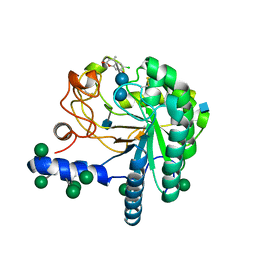 | | Hypocrea jecorina Cel6A D221A mutant soaked with 6-chloro-4- methylumbelliferyl-beta-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-7-hydroxy-4-methyl-2H-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
1QJW
 
 | | CEL6A (Y169F) WITH A NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), ... | | Authors: | Zou, J.-Y, Jones, T.A. | | Deposit date: | 1999-07-06 | | Release date: | 1999-09-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Evidence for Substrate Ring Distortion and Protein Conformational Changes During Catalysis in Cellobiohydrolase Cel6A from Trichoderma Reesei
Structure, 7, 1999
|
|
1CB2
 
 | | CELLOBIOHYDROLASE II, CATALYTIC DOMAIN, MUTANT Y169F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, alpha-D-mannopyranose | | Authors: | Kleywegt, G.J, Szardenings, M, Jones, T.A. | | Deposit date: | 1995-11-25 | | Release date: | 1996-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of Trichoderma reesei cellobiohydrolase II: the role of tyrosine 169.
Protein Eng., 9, 1996
|
|
3CBH
 
 | |
1QK2
 
 | | WILD TYPE CEL6A WITH A NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside, ... | | Authors: | Zou, J.-Y, Jones, T.A. | | Deposit date: | 1999-07-09 | | Release date: | 1999-09-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Evidence for Substrate Ring Distortion and Protein Conformational Changes During Catalysis in Cellobiohydrolase Cel6A from Trichoderma Reesei
Structure, 7, 1999
|
|
1HGW
 
 | | CEL6A D175A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), COBALT (II) ION, ... | | Authors: | Zou, J.-Y, Jones, T.A. | | Deposit date: | 2000-12-15 | | Release date: | 2002-01-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Active Site of Cellobiohydrolase Cel6A from Trichoderma Reesei: The Roles of Aspartic Acids D221 and D175
J.Am.Chem.Soc., 124, 2002
|
|
1QK0
 
 | | CEL6A WITH A NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-IODO-BENZYL ALCOHOL, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), ... | | Authors: | Zou, J.-Y, Jones, T.A. | | Deposit date: | 1999-07-08 | | Release date: | 1999-09-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Evidence for Substrate Ring Distortion and Protein Conformational Changes During Catalysis in Cellobiohydrolase Cel6A from Trichoderma Reesei
Structure, 7, 1999
|
|
1HGY
 
 | | CEL6A D221A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), alpha-D-glucopyranose, ... | | Authors: | Zou, J.-Y, Kleywegt, G.J, Jones, T.A. | | Deposit date: | 2000-12-15 | | Release date: | 2002-01-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site of Cellobiohydrolase Cel6A from Trichoderma Reesei: The Roles of Aspartic Acids D221 and D175
J.Am.Chem.Soc., 124, 2002
|
|
4AX6
 
 | | HYPOCREA JECORINA CEL6A D221A MUTANT SOAKED WITH 6-CHLORO-4- PHENYLUMBELLIFERYL-BETA-CELLOBIOSIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloranyl-7-oxidanyl-4-phenyl-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-06-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
