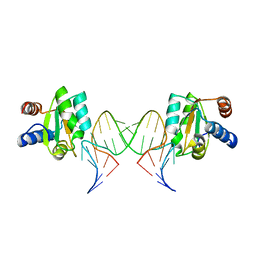
6Q8U
 
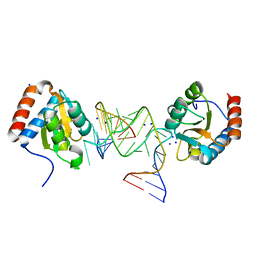 | |
4BW0
 
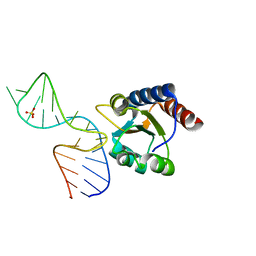 | |
5G4U
 
 | |
1RLG
 
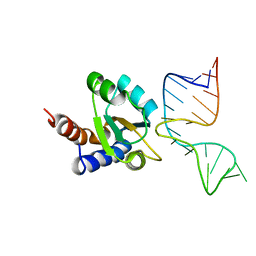 | | Molecular basis of Box C/D RNA-protein interaction: co-crystal structure of the Archaeal sRNP intiation complex | | Descriptor: | 25-MER, 50S ribosomal protein L7Ae | | Authors: | Moore, T, Zhang, Y, Fenley, M.O, Li, H. | | Deposit date: | 2003-11-25 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Basis of Box C/D RNA-Protein Interactions; Cocrystal Structure of Archaeal L7Ae and a Box C/D RNA.
STRUCTURE, 12, 2004
|
|
5G4V
 
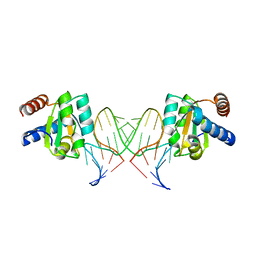 | |
5FJ4
 
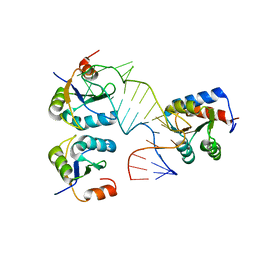 | |
4C4W
 
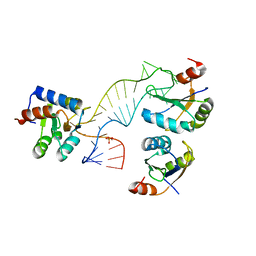 | | Structure of a rare, non-standard sequence k-turn bound by L7Ae protein | | Descriptor: | 50S RIBOSOMAL PROTEIN L7AE, DIHYDROGENPHOSPHATE ION, TSKT-23, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2013-09-09 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of a Rare, Non-Standard Sequence K-Turn Bound by L7Ae Protein
Nucleic Acids Res., 42, 2014
|
|
6HCT
 
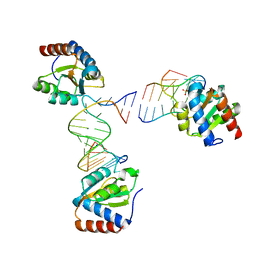 | |
