3EIF
 
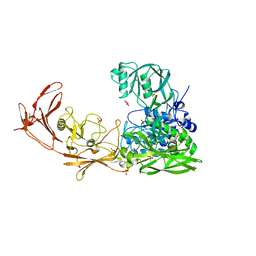 | | 1.9 angstrom crystal structure of the active form of the C5a peptidase from Streptococcus pyogenes (ScpA) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | Authors: | Cooney, J.C, Kagawa, T.F, O'Connell, M.R, Paoli, M, Mouat, P, O'Toole, P.W. | | Deposit date: | 2008-09-15 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Model for Substrate Interactions in C5a Peptidase from Streptococcus pyogenes: A 1.9 A Crystal Structure of the Active Form of ScpA
J.Mol.Biol., 386, 2009
|
|
7YZX
 
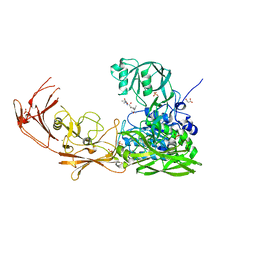 | | ScpA from Streptococcus pyogenes, D783A mutant. | | Descriptor: | C5a peptidase, CALCIUM ION, MALONIC ACID, ... | | Authors: | Kagawa, T.F, Cooney, J.C, Jain, M. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray diffraction data for the C5a-peptidase mutant with modified activity and specificity.
Data Brief, 46, 2023
|
|
7BJ3
 
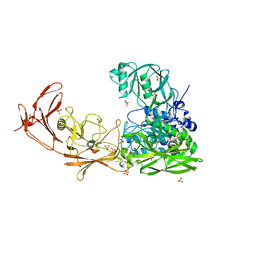 | | ScpA from Streptococcus pyogenes, S512A active site mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | Authors: | Kagawa, T.F, O'Connell, M.R, Cooney, J.C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enzyme kinetic and binding studies identify determinants of specificity for the immunomodulatory enzyme ScpA, a C5a inactivating bacterial protease.
Comput Struct Biotechnol J, 19, 2021
|
|
