4V20
 
 | | The 3-D structure of the cellobiohydrolase, Cel7A, from Aspergillus fumigatus, disaccharide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CELLOBIOHYDROLASE, ... | | Authors: | Moroz, O.V, Maranta, M, Shaghasi, T, Harris, P.V, Wilson, K.S, Davies, G.J. | | Deposit date: | 2014-10-05 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of the Cellobiohydrolase Cel7A from Aspergillus Fumigatus at 1.5 A Resolution
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1J01
 
 | | Crystal Structure Of The Xylanase Cex With Xylobiose-Derived Inhibitor Isofagomine lactam | | Descriptor: | (3S,4R)-3-hydroxy-2-oxopiperidin-4-yl beta-D-xylopyranoside, beta-1,4-xylanase | | Authors: | Williams, S.J, Notenboom, V, Wicki, J, Rose, D.R, Withers, S.G. | | Deposit date: | 2002-10-25 | | Release date: | 2002-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A New, Simple, High-Affinity Glycosidase Inhibitor: Analysis of Binding through X-ray Crystallography, Mutagenesis, and Kinetic Analysis
J.Am.Chem.Soc., 122, 2000
|
|
1Z3V
 
 | | Structure of Phanerochaete chrysosporium cellobiohydrolase Cel7D (CBH58) in complex with lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, cellulase | | Authors: | Ubhayasekera, W, Munoz, I.G, Stahlberg, J, Mowbray, S.L. | | Deposit date: | 2005-03-14 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structures of Phanerochaete chrysosporium Cel7D in complex with product and inhibitors
Febs J., 272, 2005
|
|
6GRN
 
 | | CELLOBIOHYDROLASE I (CEL7A) FROM Trichoderma reesei with S-dihydroxypropranolol in the active site | | Descriptor: | 2-[[(2~{S})-3-naphthalen-1-yloxy-2-oxidanyl-propyl]amino]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Sandgren, M, Fagerstrom, A, Widmalm, G, Stahlberg, J. | | Deposit date: | 2018-06-11 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Enantioselective Binding of Propranolol and Analogues Thereof to Cellobiohydrolase Cel7A.
Chemistry, 24, 2018
|
|
4V1Z
 
 | | The 3-D structure of the cellobiohydrolase, Cel7A, from Aspergillus fumigatus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE, ZINC ION | | Authors: | Moroz, O.V, Maranta, M, Shaghasi, T, Harris, P.V, Wilson, K.S, Davies, G.J. | | Deposit date: | 2014-10-04 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Three-Dimensional Structure of the Cellobiohydrolase Cel7A from Aspergillus Fumigatus at 1.5 A Resolution
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6CEL
 
 | | CBH1 (E212Q) CELLOPENTAOSE COMPLEX | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
4IPM
 
 | | Crystal structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with thiocellobiose | | Descriptor: | ACETATE ION, CALCIUM ION, GH7 family protein, ... | | Authors: | McGeehan, J.E, Martin, R.N.A, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2013-01-10 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2EXO
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THE BETA-1,4-GLYCANASE CEX FROM CELLULOMONAS FIMI | | Descriptor: | EXO-1,4-BETA-D-GLYCANASE | | Authors: | White, A, Withers, S.G, Gilkes, N.R, Rose, D.R. | | Deposit date: | 1994-07-11 | | Release date: | 1995-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the catalytic domain of the beta-1,4-glycanase cex from Cellulomonas fimi.
Biochemistry, 33, 1994
|
|
2HIS
 
 | | CELLULOMONAS FIMI XYLANASE/CELLULASE DOUBLE MUTANT E127A/H205N WITH COVALENT CELLOBIOSE | | Descriptor: | CELLULOMONAS FIMI FAMILY 10 BETA-1,4-GLYCANASE, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Notenboom, V, Birsan, C, Nitz, M, Rose, D.R, Warren, R.A.J, Wither, S.G. | | Deposit date: | 1998-02-23 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Insights into transition state stabilization of the beta-1,4-glycosidase Cex by covalent intermediate accumulation in active site mutants.
Nat.Struct.Biol., 5, 1998
|
|
4P1J
 
 | | Crystal structure of wild type Hypocrea jecorina Cel7a in a hexagonal crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Exoglucanase 1, SAMARIUM (III) ION, ... | | Authors: | Bodenheimer, A.B, Cuneo, M.J, Swartz, P.D, Myles, D.A, Meilleur, F. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of wild type Hypocrea jecorina Cel7a in a hexagonal crystal form
To Be Published
|
|
7CEL
 
 | | CBH1 (E217Q) IN COMPLEX WITH CELLOHEXAOSE AND CELLOBIOSE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
7NYT
 
 | | Trichoderma reesei Cel7A E212Q mutant in complex with lactose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, Exoglucanase 1, ... | | Authors: | Haataja, T, Sandgren, M, Stahlberg, J. | | Deposit date: | 2021-03-23 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: insights from crystal structures and MD simulation.
Febs J., 290, 2023
|
|
7OC8
 
 | | Trichoderma reesei Cel7A E212Q mutant in complex with pNPL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, Exoglucanase 1, ... | | Authors: | Haataja, T, Sandgren, M, Stahlberg, J. | | Deposit date: | 2021-04-26 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: insights from crystal structures and MD simulation.
Febs J., 290, 2023
|
|
7CBD
 
 | | Catalytic domain of Cellulomonas fimi Cel6B | | Descriptor: | Exoglucanase A | | Authors: | Nakamura, A, Ishiwata, D, Visootsat, A, Uchiyama, T, Mizutani, K, Kaneko, S, Murata, T, Igarashi, K, Iino, R. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Domain architecture divergence leads to functional divergence in binding and catalytic domains of bacterial and fungal cellobiohydrolases.
J.Biol.Chem., 295, 2020
|
|
2MWK
 
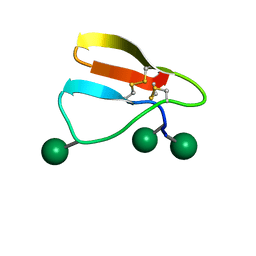 | | Family 1 Carbohydrate-Binding Module from Trichoderma reesei Cel7A with O-mannose residues at Thr1, Ser3, and Ser14 | | Descriptor: | Exoglucanase 1, alpha-D-mannopyranose | | Authors: | Happs, R.M, Chen, L, Resch, M.G, Davis, M.F, Beckham, G.T, Tan, Z, Crowley, M.F. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | O-glycosylation effects on family 1 carbohydrate-binding module solution structures.
Febs J., 282, 2015
|
|
2MWJ
 
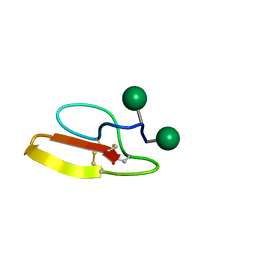 | | Solution structure of Family 1 Carbohydrate-Binding Module from Trichoderma reesei Cel7A with O-mannose residues at Thr1 and Ser3 | | Descriptor: | Exoglucanase 1, alpha-D-mannopyranose | | Authors: | Happs, R.M, Chen, L, Resch, M.G, Davis, M.F, Beckham, G.T, Tan, Z, Crowley, M.F. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | O-glycosylation effects on family 1 carbohydrate-binding module solution structures.
Febs J., 282, 2015
|
|
4P1H
 
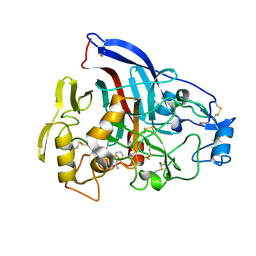 | | Crystal structure of wild type Hypocrea jecorina Cel7a in a monoclinic crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, Exoglucanase 1, ... | | Authors: | Bodenheimer, A.B, Cuneo, M.J, Swartz, P.D, Myles, D.A, Meilleur, F. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of wild type Hypocrea jecorina Cel7a in a monoclinic crystal form
to be published
|
|
4JJJ
 
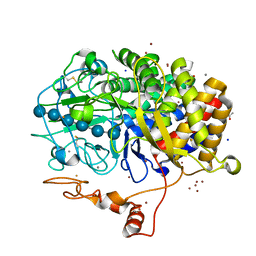 | | The structure of T. fusca GH48 D224N mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2013-03-07 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cel48A from Thermobifida fusca: structure and site directed mutagenesis of key residues.
Biotechnol.Bioeng., 111, 2014
|
|
5X39
 
 | |
5X34
 
 | |
5BV9
 
 | |
5CEL
 
 | | CBH1 (E212Q) CELLOTETRAOSE COMPLEX | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
1Z3W
 
 | | Structure of Phanerochaete chrysosporium cellobiohydrolase Cel7D (CBH58) in complex with cellobioimidazole | | Descriptor: | (5R,6R,7R,8S)-7,8-dihydroxy-5-(hydroxymethyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-6-yl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, cellulase | | Authors: | Ubhayasekera, W, Vasella, A, Stahlberg, J, Mowbray, S.L. | | Deposit date: | 2005-03-14 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Phanerochaete chrysosporium Cel7D in complex with product and inhibitors
Febs J., 272, 2005
|
|
1Z3T
 
 | |
1Q2B
 
 | | CELLOBIOHYDROLASE CEL7A WITH DISULPHIDE BRIDGE ADDED ACROSS EXO-LOOP BY MUTATIONS D241C AND D249C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, EXOCELLOBIOHYDROLASE I | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D.
J.Mol.Biol., 333, 2003
|
|
