9F0X
 
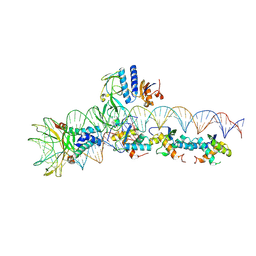 | | CryoEM structure of the F plasmid relaxosome in its pre-initiation state, derived from the ds-27_+143-R Locally-refined Map 3.76 A | | Descriptor: | Integration host factor subunit alpha, Integration host factor subunit beta, Multifunctional conjugation protein TraI, ... | | Authors: | Williams, S.M, Waksman, G. | | Deposit date: | 2024-04-17 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Cryo-EM Structure of the relaxosome, a complex essential for bacterial mating and the spread of antibiotic resistance genes.
Nat Commun, 16, 2025
|
|
9F0Z
 
 | | CryoEM structure of the F plasmid relaxosome with truncated TraI1-863 in its TE mode, derived from the ss-27_+8ds+9_+143-R_deltaAH+CTD Locally-refined 3.42 A Map | | Descriptor: | Integration host factor subunit alpha, Integration host factor subunit beta, MAGNESIUM ION, ... | | Authors: | Williams, S.M, Waksman, G. | | Deposit date: | 2024-04-17 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Cryo-EM Structure of the relaxosome, a complex essential for bacterial mating and the spread of antibiotic resistance genes.
Nat Commun, 16, 2025
|
|
9F12
 
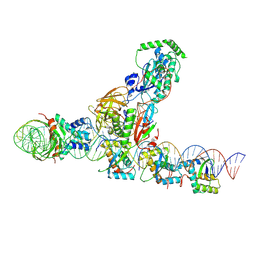 | | CryoEM structure of the F plasmid relaxosome with oriT DNA ss-27_-3ds-2_+143 and TraI its TE mode, derived from ss-27_-3ds-2_+143-R Locally-refined 3.42 A Map. | | Descriptor: | Integration host factor subunit alpha, Integration host factor subunit beta, MAGNESIUM ION, ... | | Authors: | Williams, S.M, Waksman, G. | | Deposit date: | 2024-04-17 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Cryo-EM Structure of the relaxosome, a complex essential for bacterial mating and the spread of antibiotic resistance genes.
Nat Commun, 16, 2025
|
|
9F11
 
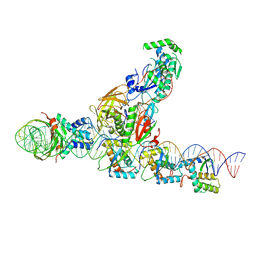 | | CryoEM structure of the F plasmid relaxosome with oriT DNA ss-27_+3ds+4_+143 and TraI its TE mode, derived from ss-27_+3ds+4_+143-R Locally-refined 3.68 A Map. | | Descriptor: | Integration host factor subunit alpha, Integration host factor subunit beta, MAGNESIUM ION, ... | | Authors: | Williams, S.M, Waksman, G. | | Deposit date: | 2024-04-17 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Cryo-EM Structure of the relaxosome, a complex essential for bacterial mating and the spread of antibiotic resistance genes.
Nat Commun, 16, 2025
|
|
9F0Y
 
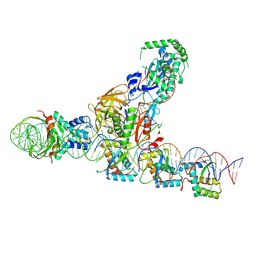 | | CryoEM structure of the F plasmid relaxosome with TraI in its TE mode, derived from the ss-27_+8ds+9_+143-R Locally-refined 3.45 A Map. | | Descriptor: | Integration host factor subunit alpha, Integration host factor subunit beta, MAGNESIUM ION, ... | | Authors: | Williams, S.M, Waksman, G. | | Deposit date: | 2024-04-17 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM Structure of the relaxosome, a complex essential for bacterial mating and the spread of antibiotic resistance genes.
Nat Commun, 16, 2025
|
|
9F10
 
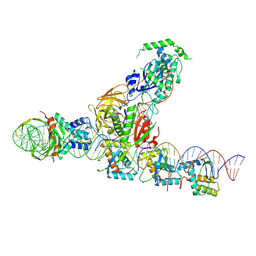 | | CryoEM structure of the F plasmid relaxosome with TraI in its TE mode, without accessory protein TraM. Derived from the ss-27_+8ds+9_+143-R_deltaTraM Locally-refined 2.94 A Map. | | Descriptor: | Integration host factor subunit alpha, Integration host factor subunit beta, MAGNESIUM ION, ... | | Authors: | Williams, S.M, Waksman, G. | | Deposit date: | 2024-04-17 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM Structure of the relaxosome, a complex essential for bacterial mating and the spread of antibiotic resistance genes.
Nat Commun, 16, 2025
|
|
