1FAE
 
 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellobiose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-13 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1F9O
 
 | | Crystal structure of the cellulase Cel48F from C. Cellulolyticum with the thiooligosaccharide inhibitor PIPS-IG3 | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-iodophenyl 1,4-dithio-beta-D-glucopyranoside | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-11 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1FBO
 
 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellobiitol | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-D-glucose | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-16 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1F9D
 
 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellotetraose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-10 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1FBW
 
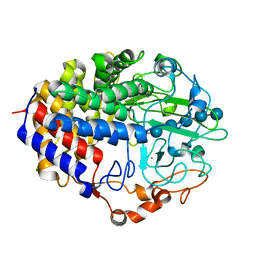 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellohexaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1G9G
 
 | | XTAL-STRUCTURE OF THE FREE NATIVE CELLULASE CEL48F | | Descriptor: | CALCIUM ION, CELLULASE CEL48F, MAGNESIUM ION | | Authors: | Parsiegla, G, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-11-23 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of mutants of cellulase Cel48F of Clostridium cellulolyticum in complex with long hemithiocellooligosaccharides give rise to a new view of the substrate pathway during processive action
J.Mol.Biol., 375, 2008
|
|
1G9J
 
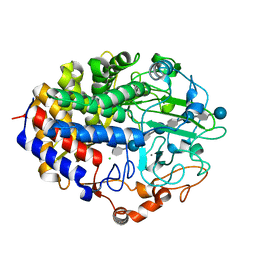 | | X-TAL STRUCTURE OF THE MUTANT E44Q OF THE CELLULASE CEL48F IN COMPLEX WITH A THIOOLIGOSACCHARIDE | | Descriptor: | CALCIUM ION, CELLULASE CEL48F, CHLORIDE ION, ... | | Authors: | Parsiegla, G, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-11-24 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of mutants of cellulase Cel48F of Clostridium cellulolyticum in complex with long hemithiocellooligosaccharides give rise to a new view of the substrate pathway during processive action
J.Mol.Biol., 375, 2008
|
|
