1J9B
 
 | | ARSENATE REDUCTASE+0.4M ARSENITE FROM E. COLI | | Descriptor: | ARSENATE REDUCTASE, CESIUM ION, SULFATE ION, ... | | Authors: | Martin, P, Edwards, B.F. | | Deposit date: | 2001-05-24 | | Release date: | 2001-12-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme.
Structure, 9, 2001
|
|
1J9C
 
 | | Crystal Structure of tissue factor-factor VIIa complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-phenylalaninamide, ... | | Authors: | Huang, M, Ruf, W, Edgington, T.S, Wilson, I.A. | | Deposit date: | 2001-05-24 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand Induced Conformational Transitions of Tissue Factor. Crystal Structure of the Tissue Factor:Factor VIIa Complex.
To be Published
|
|
1J9E
 
 | | Low Temperature (100K) Crystal Structure of Flavodoxin D. vulgaris S35C Mutant at 1.44 Angstrom Resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Artali, R, Bombieri, G, Meneghetti, F, Gilardi, G, Sadeghi, S.J, Cavazzini, D, Rossi, G.L. | | Deposit date: | 2001-05-25 | | Release date: | 2001-09-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Comparison of the refined crystal structures of wild-type (1.34 A) flavodoxin from Desulfovibrio vulgaris and the S35C mutant (1.44 A) at 100 K.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1J9G
 
 | | Low Temperature (100K) Crystal Structure of Flavodoxin D. vulgaris S64C Mutant, monomer oxidised, at 2.4 Angstrom Resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Artali, R, Bombieri, G, Meneghetti, F, Gilardi, G, Sadeghi, S.J, Cavazzini, D, Rossi, G.L. | | Deposit date: | 2001-05-25 | | Release date: | 2001-09-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparison of the refined crystal structures of wild-type (1.34 A) flavodoxin from Desulfovibrio vulgaris and the S35C mutant (1.44 A) at 100 K.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1J9H
 
 | | Crystal Structure of an RNA Duplex with Uridine Bulges | | Descriptor: | 5'-R(*GP*UP*GP*UP*CP*GP*(CBR)P*AP*C)-3', CALCIUM ION | | Authors: | Xiong, Y, Deng, J, Sudarsanakumar, C, Sundaralingam, M. | | Deposit date: | 2001-05-25 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an RNA duplex r(gugucgcac)(2) with uridine bulges.
J.Mol.Biol., 313, 2001
|
|
1J9I
 
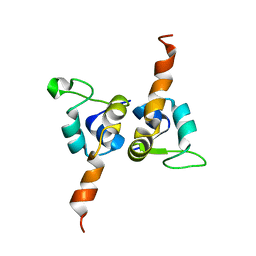 | | STRUCTURE OF THE DNA BINDING DOMAIN OF THE GPNU1 SUBUNIT OF LAMBDA TERMINASE | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | De Beer, T, Meyer, J, Ortega, M, Yang, Q, Maes, L, Duffy, C, Berton, N, Sippy, J, Overduin, M, Feiss, M, Catalano, C. | | Deposit date: | 2001-05-25 | | Release date: | 2002-08-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Insights into specific DNA recognition during the assembly of a viral genome packaging machine.
Mol.Cell, 9, 2002
|
|
1J9J
 
 | | CRYSTAL STRUCTURE ANALYSIS OF SURE PROTEIN FROM T.MARITIMA | | Descriptor: | MAGNESIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, SULFATE ION | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-27 | | Release date: | 2001-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
1J9K
 
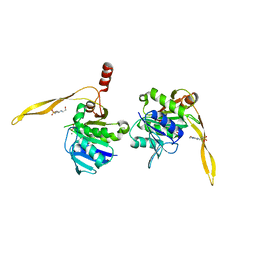 | | CRYSTAL STRUCTURE OF SURE PROTEIN FROM T.MARITIMA IN COMPLEX WITH TUNGSTATE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, ... | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-27 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
1J9L
 
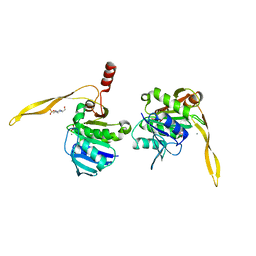 | | CRYSTAL STRUCTURE OF SURE PROTEIN FROM T.MARITIMA IN COMPLEX WITH VANADATE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, ... | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-28 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
1J9M
 
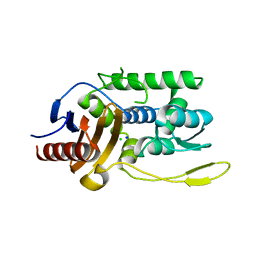 | | K38H mutant of Streptomyces K15 DD-transpeptidase | | Descriptor: | CHLORIDE ION, DD-transpeptidase, SODIUM ION | | Authors: | Fonze, E, Rhazi, N, Nguyen-Disteche, M, Charlier, P. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1J9N
 
 | | Solution Structure of the Nucleopeptide [AC-LYS-TRP-LYS-HSE(p3*dGCATCG)-ALA]-[p5*dCGTAGC] | | Descriptor: | 5'-D(*CP*GP*TP*AP*GP*C)-3', 5'-D(*GP*CP*TP*AP*CP*(PGN))-3', peptide ACE-LYS-TRP-LYS-HSE-ALA | | Authors: | Gomez-Pinto, I, Marchan, V, Gago, F, Grandas, A, Gonzalez, C. | | Deposit date: | 2001-05-28 | | Release date: | 2003-01-21 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability of tryptophan-containing nucleopeptide duplexes
Chembiochem, 4, 2003
|
|
1J9O
 
 | | SOLUTION STRUCTURE OF HUMAN LYMPHOTACTIN | | Descriptor: | LYMPHOTACTIN | | Authors: | Kuloglu, E.S, McCaslin, D.R, Kitabwalla, M, Pauza, C.D, Markley, J.L, Volkman, B.F. | | Deposit date: | 2001-05-28 | | Release date: | 2001-10-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Monomeric solution structure of the prototypical 'C' chemokine lymphotactin.
Biochemistry, 40, 2001
|
|
1J9Q
 
 | | Crystal structure of nitrite soaked oxidized D98N AFNIR | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION | | Authors: | Boulanger, M.J, Murphy, M.E. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Alternate substrate binding modes to two mutant (D98N and H255N) forms of nitrite reductase from Alcaligenes faecalis S-6: structural model of a transient catalytic intermediate
Biochemistry, 40, 2001
|
|
1J9R
 
 | | Crystal structure of nitrite soaked reduced D98N AFNIR | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION | | Authors: | Boulanger, M.J, Murphy, M.E. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Alternate substrate binding modes to two mutant (D98N and H255N) forms of nitrite reductase from Alcaligenes faecalis S-6: structural model of a transient catalytic intermediate
Biochemistry, 40, 2001
|
|
1J9S
 
 | | Crystal structure of nitrite soaked oxidized H255N AFNIR | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION | | Authors: | Boulanger, M.J, Murphy, M.E. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternate substrate binding modes to two mutant (D98N and H255N) forms of nitrite reductase from Alcaligenes faecalis S-6: structural model of a transient catalytic intermediate
Biochemistry, 40, 2001
|
|
1J9T
 
 | | Crystal structure of nitrite soaked reduced H255N AFNIR | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION | | Authors: | Boulanger, M.J, Murphy, M.E. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Alternate substrate binding modes to two mutant (D98N and H255N) forms of nitrite reductase from Alcaligenes faecalis S-6: structural model of a transient catalytic intermediate
Biochemistry, 40, 2001
|
|
1J9V
 
 | | Solution structure of a lactam analogue (DabD) of HIV gp41 600-612 loop. | | Descriptor: | DabD (Ace)IWG(DAB)SGKLIDTTA ANALOGUE OF HIV GP41 | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-05-29 | | Release date: | 2003-07-01 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
1J9W
 
 | | Solution Structure of the CAI Michigan 1 Variant | | Descriptor: | 1,2-ETHANEDIOL, CARBONIC ANHYDRASE I, ZINC ION | | Authors: | Briganti, F, Ferraroni, M, Chedwiggen, W.R, Scozzafava, A, Supuran, C.T, Tilli, S. | | Deposit date: | 2001-05-29 | | Release date: | 2001-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a zinc-activated variant of human carbonic anhydrase I, CA I Michigan 1: evidence for a second zinc binding site involving arginine coordination.
Biochemistry, 41, 2002
|
|
1J9Y
 
 | | Crystal structure of mannanase 26A from Pseudomonas cellulosa | | Descriptor: | MANNANASE A, ZINC ION | | Authors: | Hogg, D, Woo, E.-J, Bolam, D.N, McKie, V.A, Gilbert, H.J, Pickersgill, R.W. | | Deposit date: | 2001-05-29 | | Release date: | 2001-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of mannanase 26A from Pseudomonas cellulosa and analysis of residues involved in substrate binding
J.Biol.Chem., 276, 2001
|
|
1J9Z
 
 | | CYPOR-W677G | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hubbard, P.A, Shen, A.L, Paschke, R, Kasper, C.B, Kim, J.J. | | Deposit date: | 2001-05-29 | | Release date: | 2001-08-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | NADPH-cytochrome P450 oxidoreductase. Structural basis for hydride and electron transfer.
J.Biol.Chem., 276, 2001
|
|
1JA0
 
 | | CYPOR-W677X | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hubbard, P.A, Shen, A.L, Paschke, R, Kasper, C.B, Kim, J.J. | | Deposit date: | 2001-05-29 | | Release date: | 2001-08-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | NADPH-cytochrome P450 oxidoreductase. Structural basis for hydride and electron transfer.
J.Biol.Chem., 276, 2001
|
|
1JA1
 
 | | CYPOR-Triple Mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hubbard, P.A, Shen, A.L, Paschke, R, Kasper, C.B, Kim, J.J. | | Deposit date: | 2001-05-29 | | Release date: | 2001-08-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NADPH-cytochrome P450 oxidoreductase. Structural basis for hydride and electron transfer.
J.Biol.Chem., 276, 2001
|
|
1JA2
 
 | |
1JA3
 
 | | Crystal Structure of the Murine NK Cell Inhibitory Receptor Ly-49I | | Descriptor: | MHC class I recognition receptor Ly49I | | Authors: | Dimasi, N, Sawicki, W.M, Reineck, L.A, Li, Y, Natarajan, K, Murgulies, D.H, Mariuzza, A.R. | | Deposit date: | 2001-05-29 | | Release date: | 2002-07-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Ly49I natural killer cell receptor reveals variability in dimerization mode within the Ly49 family.
J.Mol.Biol., 320, 2002
|
|
1JA4
 
 | |
