4DN4
 
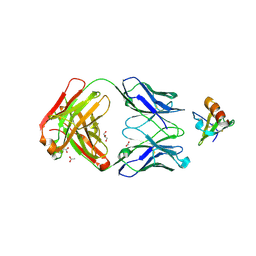 | | Crystal structure of the complex between cnto888 fab and mcp-1 mutant p8a | | Descriptor: | ACETATE ION, C-C motif chemokine 2, CNTO888 HEAVY CHAIN, ... | | Authors: | Obmolova, G, Teplyakov, A, Malia, T, Grygiel, T, Sweet, R, Snyder, L, Gilliland, G. | | Deposit date: | 2012-02-08 | | Release date: | 2012-10-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for high selectivity of anti-CCL2 neutralizing antibody CNTO 888.
Mol.Immunol., 51, 2012
|
|
3TN2
 
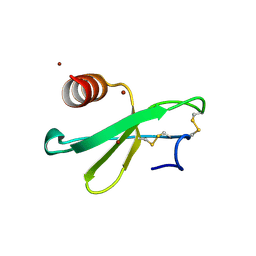 | | structure analysis of MIP1-beta P8A | | Descriptor: | C-C motif chemokine 4, ZINC ION | | Authors: | Guo, Q, Tang, W.J. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-05 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
6FGP
 
 | |
6N2U
 
 | |
1ZXT
 
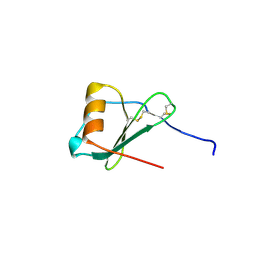 | | Crystal Structure of A Viral Chemokine | | Descriptor: | functional macrophage inflammatory protein 1-alpha homolog | | Authors: | Luz, J.G, Yu, M, Su, Y, Wu, Z, Zhou, Z, Sun, R, Wilson, I.A. | | Deposit date: | 2005-06-08 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of viral macrophage inflammatory protein I encoded by Kaposi's sarcoma-associated herpesvirus at 1.7A.
J.Mol.Biol., 352, 2005
|
|
5OB5
 
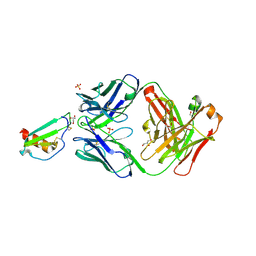 | |
6SHR
 
 | | X-RAY CRYSTAL STRUCTURE OF CELL-FREE PROTEIN SYNTHESIS (CFPS) PRODUCED SDF1-A | | Descriptor: | Stromal cell-derived factor 1 | | Authors: | Jugnarain, V.M, Mitchell, E, Forsyth, T, Michael, H, Cortes, S, Tillier, B. | | Deposit date: | 2019-08-08 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.745 Å) | | Cite: | X-RAY CRYSTAL STRUCTURE OF CELL-FREE PROTEIN SYNTHESIS (CFPS) PRODUCED SDF1-A
To Be Published
|
|
3KBX
 
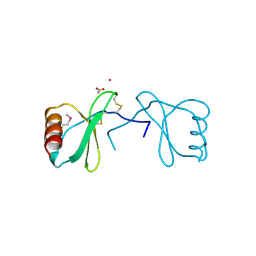 | |
6STK
 
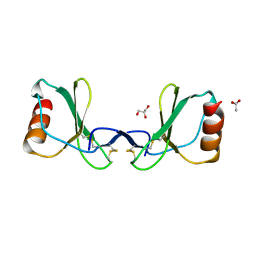 | |
1HA6
 
 | | NMR Solution Structure of Murine CCL20/MIP-3a Chemokine | | Descriptor: | MACROPHAGE INFLAMMATORY PROTEIN 3 ALPHA | | Authors: | Perez-Canadillas, J.M, Zaballos, A, Gutierrez, J, Varona, R, Roncal, F, Albar, J.P, Marquez, G, Bruix, M. | | Deposit date: | 2001-03-28 | | Release date: | 2001-08-22 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Murine Ccl20/Mip3-A, a Chemiokine that Specifically Chemoattracs Immature Dendritic Cells and Lymphocytes Through its Highly Specific Interaction with the Beta-Chemokine Receptor Ccr6
J.Biol.Chem., 276, 2001
|
|
1HFN
 
 | | NMR solution structures of vMIP-II 1-71 from Kaposi's sarcoma-associated herpesvirus. | | Descriptor: | VIRAL MACROPHAGE INFLAMMATORY PROTEIN-II | | Authors: | Crump, M.P, Elisseeva, E, Gong, J.-H, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 2000-12-07 | | Release date: | 2001-01-07 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure/Function of Human Herpesvirus-8 Mip-II (1-71) and the Antagonist N-Terminal Segment (1-10)
FEBS Lett., 489, 2001
|
|
5YAM
 
 | |
1VMP
 
 | | STRUCTURE OF THE ANTI-HIV CHEMOKINE VMIP-II | | Descriptor: | PROTEIN (ANTI-HIV CHEMOKINE MIP VII) | | Authors: | Liwang, A.C, Wang, Z.-X, Sun, Y, Peiper, S.C, Liwang, P.J. | | Deposit date: | 1999-03-25 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the anti-HIV chemokine vMIP-II.
Protein Sci., 8, 1999
|
|
1VMC
 
 | | STROMA CELL-DERIVED FACTOR-1ALPHA (SDF-1ALPHA) | | Descriptor: | Stromal cell-derived factor 1 | | Authors: | Gozansky, E.K, Clore, G.M. | | Deposit date: | 2004-09-20 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Mapping the binding of the N-terminal extracellular tail of the CXCR4 receptor to stromal cell-derived factor-1alpha.
J.Mol.Biol., 345, 2005
|
|
4OIJ
 
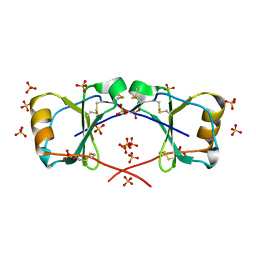 | | X-ray crystal structure of racemic non-glycosylated chemokine Ser-CCL1 | | Descriptor: | C-C motif chemokine 1, D-Ser-CCL1, SULFATE ION | | Authors: | Okamoto, R, Mandal, K, Sawaya, M.R, Kajihara, Y, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2014-01-19 | | Release date: | 2014-05-07 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (Quasi-)Racemic X-ray Structures of Glycosylated and Non-Glycosylated Forms of the Chemokine Ser-CCL1 Prepared by Total Chemical Synthesis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4OIK
 
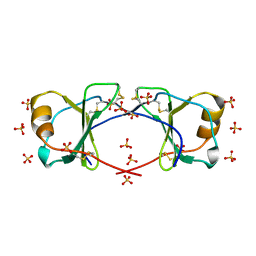 | | (Quasi-)Racemic X-ray crystal structure of glycosylated chemokine Ser-CCL1. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 1, CITRIC ACID, ... | | Authors: | Okamoto, R, Mandal, K, Sawaya, M.R, Kajihara, Y, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2014-01-19 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | (Quasi-)Racemic X-ray Structures of Glycosylated and Non-Glycosylated Forms of the Chemokine Ser-CCL1 Prepared by Total Chemical Synthesis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6AEZ
 
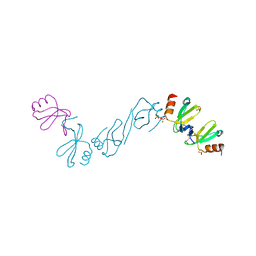 | | Crystal structure of human CCL5 trimer | | Descriptor: | C-C motif chemokine 5, SULFATE ION | | Authors: | Chen, Y.C, Li, K.M, Chen, P.J, Zarivach, R, Sun, Y.J, Sue, S.C. | | Deposit date: | 2018-08-07 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Integrative Model to Coordinate the Oligomerization and Aggregation Mechanisms of CCL5.
J.Mol.Biol., 432, 2020
|
|
3N52
 
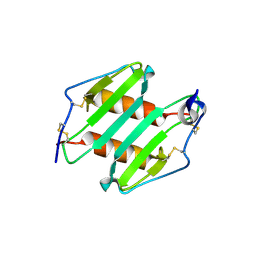 | | crystal Structure analysis of MIP2 | | Descriptor: | C-X-C motif chemokine 2 | | Authors: | Rajasekaran, D. | | Deposit date: | 2010-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Model of GAG/MIP-2/CXCR2 Interfaces and Its Functional Effects.
Biochemistry, 51, 2012
|
|
6VGJ
 
 | | N-terminal variant of CXCL13 | | Descriptor: | C-X-C motif chemokine 13 | | Authors: | Rosenberg Jr, E.M, Lolis, E.J. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The N-terminal length and side-chain composition of CXCL13 affect crystallization, structure and functional activity.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4LMQ
 
 | | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12 | | Descriptor: | Stromal cell-derived factor 1, hu30D8 Fab heavy chain, hu30D8 Fab light chain | | Authors: | Zhong, Z, Wang, J, Li, B, Xiang, H, Ultsch, M, Coons, M, Wong, T, Chiang, N.Y, Clark, S, Clark, R, Quintana, L, Gribling, P, Suto, E, Barck, K, Corpuz, R, Yao, J, Takkar, R, Lee, W.P, Damico-Beyer, L.A, Carano, R.D, Adams, C, Kelley, R.F, Wang, W, Ferrara, N. | | Deposit date: | 2013-07-10 | | Release date: | 2013-08-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12.
Clin.Cancer Res., 19, 2013
|
|
4MHE
 
 | | Crystal structure of CC-chemokine 18 | | Descriptor: | ACETATE ION, C-C motif chemokine 18 | | Authors: | Liang, W.G, Tang, W.-J. | | Deposit date: | 2013-08-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
1QE6
 
 | | INTERLEUKIN-8 WITH AN ADDED DISULFIDE BETWEEN RESIDUES 5 AND 33 (L5C/H33C) | | Descriptor: | INTERLEUKIN-8 VARIANT, SULFATE ION | | Authors: | Gerber, N, Lowman, H, Artis, D.R, Eigenbrot, C. | | Deposit date: | 1999-07-13 | | Release date: | 2000-03-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Receptor-binding conformation of the "ELR" motif of IL-8: X-ray structure of the L5C/H33C variant at 2.35 A resolution.
Proteins, 38, 2000
|
|
1QG7
 
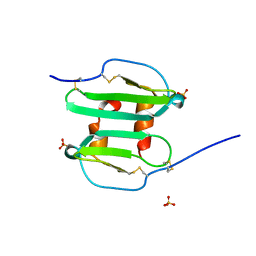 | | STROMA CELL-DERIVED FACTOR-1ALPHA (SDF-1ALPHA) | | Descriptor: | STROMAL CELL-DERIVED FACTOR 1 ALPHA, SULFATE ION | | Authors: | Senda, T, Nandhagopal, N, Sugimoto, K, Mitsui, Y. | | Deposit date: | 1999-04-21 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of recombinant native SDF-1alpha with additional mutagenesis studies: an attempt at a more comprehensive interpretation of accumulated structure-activity relationship data.
J.Interferon Cytokine Res., 20, 2000
|
|
1QNK
 
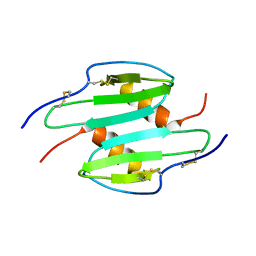 | | TRUNCATED HUMAN GROB[5-73], NMR, 20 STRUCTURES | | Descriptor: | C-X-C motif chemokine 2 | | Authors: | Qian, Y.Q, Johanson, K, McDevitt, P. | | Deposit date: | 1999-10-18 | | Release date: | 2000-02-04 | | Last modified: | 2018-06-13 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of truncated human GRObeta [5-73] and its structural comparison with CXC chemokine family members GROalpha and IL-8.
J. Mol. Biol., 294, 1999
|
|
6CWS
 
 | |
