7N2C
 
 | | Elongating 70S ribosome complex in a fusidic acid-stalled intermediate state of translocation bound to EF-G(GDP) (INT2) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-14 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
5A65
 
 | | Crystal structure of mouse thiamine triphosphatase in complex with thiamine diphosphate, orthophosphate and magnesium ions. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
7ZEG
 
 | | Human cytosolic 5' nucleotidase IIIB in complex with 3,4-diF-Bn7GMP | | Descriptor: | 1,2-ETHANEDIOL, 7-methylguanosine phosphate-specific 5'-nucleotidase, MAGNESIUM ION, ... | | Authors: | Kubacka, D, Kozarski, M, Baranowski, M.R, Wojcik, R, Jemielity, J, Kowalska, J, Basquin, J. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Substrate-Based Design of Cytosolic Nucleotidase IIIB Inhibitors and Structural Insights into Inhibition Mechanism.
Pharmaceuticals, 15, 2022
|
|
5J57
 
 | | V5E1-RTA complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ricin, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2016-04-01 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of Single Domain Antibodies Bound to a Second Neutralizing Hot Spot on Ricin Toxin's Enzymatic Subunit.
J. Biol. Chem., 292, 2017
|
|
5CVP
 
 | | Structure of Xoo1075, a peptide deformylase from Xanthomonas oryzae pv oryze, in complex with fragment 571 | | Descriptor: | 1-(4-chlorophenyl)-N-methylmethanesulfonamide, ACETATE ION, CADMIUM ION, ... | | Authors: | Ngo, H.P.T, Kang, L.W. | | Deposit date: | 2015-07-27 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Xoo1075, a peptide deformylase from Xanthomonas oryzae pv oryze, in complex with fragment 571
To Be Published
|
|
6R6Y
 
 | | Crystal structure of human carbonic anhydrase isozyme XII with 4-chloro-2-cyclohexylsulfanyl-N-(2-hydroxyethyl)-5-sulfamoyl-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-cyclohexylsulfanyl-~{N}-(2-hydroxyethyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Halogenated and di-substituted benzenesulfonamides as selective inhibitors of carbonic anhydrase isoforms.
Eur.J.Med.Chem., 185, 2020
|
|
8G9X
 
 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Changela, A, Gorman, J, Kwong, P.D. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.46 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
7N31
 
 | | Elongating 70S ribosome complex in a post-translocation (POST) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-14 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
5J74
 
 | | Fluorogen activating protein AM2.2 in complex with TO1-2p | | Descriptor: | 1-(17-amino-5,8-dioxo-12,15-dioxa-4,9-diazaheptadecan-1-yl)-4-{[3-(3-sulfopropyl)-1,3-benzothiazol-3-ium-2-yl]methyl}quinolin-1-ium, PHOSPHATE ION, scFv AM2.2 | | Authors: | Stanfield, R.L, Wilson, I.A, Wu, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Small-Molecule Nonfluorescent Inhibitors of Fluorogen-Fluorogen Activating Protein Binding Pair.
J Biomol Screen, 21, 2016
|
|
6R30
 
 | |
5J3L
 
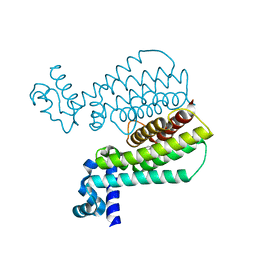 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with 1-((2-cyclopentylethyl)sulfonyl)pyrrolidine at 1.66A resolution | | Descriptor: | 1-[(2-cyclopentylethyl)sulfonyl]pyrrolidine, HTH-type transcriptional regulator EthR | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
7QQQ
 
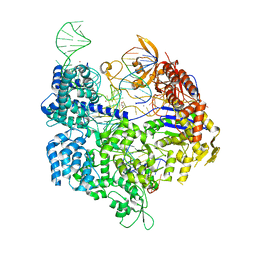 | | SpCas9 bound to AAVS1 off-target4 DNA substrate | | Descriptor: | 1,2-ETHANEDIOL, AAVS1 off-target4 non-target strand, AAVS1 off-target4 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
3JVL
 
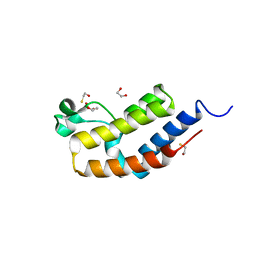 | | Crystal structure of bromodomain 2 of mouse Brd4 | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Bromodomain-containing protein 4 | | Authors: | Vollmuth, F, Blankenfeldt, W, Geyer, M. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of the Dual Bromodomains of the P-TEFb-activating Protein Brd4 at Atomic Resolution
J.Biol.Chem., 284, 2009
|
|
6V7F
 
 | | Human Arginase1 Complexed with Bicyclic Inhibitor Compound 13 | | Descriptor: | Arginase-1, MANGANESE (II) ION, {3-[(5R,7S,8S)-8-azaniumyl-8-carboxy-2-azaspiro[4.4]nonan-2-ium-7-yl]propyl}(trihydroxy)borate(1-) | | Authors: | Palte, R.L, Lesburg, C.A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery and Optimization of Rationally Designed Bicyclic Inhibitors of Human Arginase to Enhance Cancer Immunotherapy.
Acs Med.Chem.Lett., 11, 2020
|
|
6R8R
 
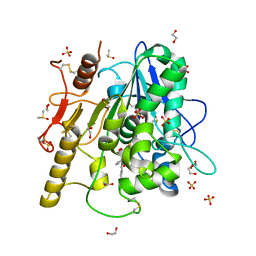 | | Structure of the Wnt deacylase Notum in complex with isoquinoline 45 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Vecchia, L, Zhao, Y, Ruza, R.R, Jones, E.Y. | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Discovery of 2-phenoxyacetamides as inhibitors of the Wnt-depalmitoleating enzyme NOTUM from an X-ray fragment screen.
Medchemcomm, 10, 2019
|
|
7CAJ
 
 | | Crystal structure of SETDB1 Tudor domain in complexed with Compound 2. | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-5H-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Guo, Y.P, Liang, X, Xin, M, Luyi, H, Chengyong, W, Yang, S.Y. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6I14
 
 | | CRYSTAL STRUCTURE OF FASCIN IN COMPLEX WITH COMPOUND 9 | | Descriptor: | 2-[(3,4-dichlorophenyl)methyl]-~{N}-(1-methylpyrazol-4-yl)-1-oxidanylidene-isoquinoline-4-carboxamide, ACETATE ION, Fascin | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2018-10-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-based design, synthesis and biological evaluation of a novel series of isoquinolone and pyrazolo[4,3-c]pyridine inhibitors of fascin 1 as potential anti-metastatic agents.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5YRX
 
 | | Crystal structure of a hypothetical protein Rv3716c from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Nucleoid-associated protein Rv3716c | | Authors: | Deka, G, Gopalan, A, Prabhavathi, M, Savithri, H.S, Raja, A, Murthy, M.R.N. | | Deposit date: | 2017-11-10 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biophysical characterization of Rv3716c, a hypothetical protein from Mycobacterium tuberculosis
Biochem. Biophys. Res. Commun., 495, 2018
|
|
7USH
 
 | | BRD2-BD2 in complex with SF2523 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-(morpholin-4-yl)-7H-thieno[3,2-b]pyran-7-one, Bromodomain-containing protein 2 | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
8ATH
 
 | | CRYSTAL STRUCTURE OF LAMP1 IN COMPLEX WITH FAB-B. | | Descriptor: | Fab B Heavy Chain, Fab B Light Chain, Lysosome-associated membrane glycoprotein 1 | | Authors: | Mathieu, M, Dupuy, A. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.366 Å) | | Cite: | Deciphering cross-species reactivity of LAMP-1 antibodies using deep mutational epitope mapping and AlphaFold.
Mabs, 15, 2023
|
|
6BSU
 
 | | Crystal structure of xyloglucan xylosyltransferase I | | Descriptor: | MANGANESE (II) ION, Xyloglucan 6-xylosyltransferase 1 | | Authors: | Culbertson, A.T, Ehrlich, J.J, Choe, J, Honzatko, R.B, Zabotina, O.A. | | Deposit date: | 2017-12-04 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5I3E
 
 | |
3JZ6
 
 | | Crystal structure of Mycobacterium smegmatis Branched Chain Aminotransferase in complex with pyridoxal-5'-phosphate at 1.9 angstrom. | | Descriptor: | Branched-chain amino acid aminotransferase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Castell, A, Mille, C, Unge, T. | | Deposit date: | 2009-09-23 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of mycobacterial branched-chain aminotransferase: implications for inhibitor design.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6Z3I
 
 | | Structure of recombinant beta-glucocerebrosidase in complex with bifunctional cyclophellitol aziridine activity based probe | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosomal acid glucosylceramidase, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-05-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
6BYA
 
 | | Crystal structure of LdBPK_091320 with inhibitor bound | | Descriptor: | 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, UNKNOWN ATOM OR ION, Uncharacterized protein | | Authors: | Dong, A, Lin, Y.H, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of LdBPK_091320.1 with with inhibitor bound
to be published
|
|
