5MXW
 
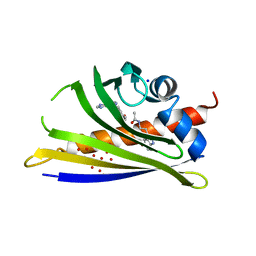 | | Crystal structure of yellow lupin LLPR-10.2B protein in complex with melatonin and trans-zeatin. | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, Class 10 plant pathogenesis-related protein, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Sliwiak, J, Sikorski, M, Jaskolski, M. | | Deposit date: | 2017-01-25 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PR-10 proteins as potential mediators of melatonin-cytokinin cross-talk in plants: crystallographic studies of LlPR-10.2B isoform from yellow lupine.
FEBS J., 285, 2018
|
|
9BOI
 
 | | Cryo-EM structure of human Spns1 in complex with LPC (18:1) | | Descriptor: | (4R,7R,18Z)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, mVenus,Maltose/maltodextrin-binding periplasmic protein,Protein spinster homolog 1,Designed ankyrin repeat protein (DARPin) | | Authors: | Chen, H, Li, X. | | Deposit date: | 2024-05-03 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular basis of Spns1-mediated lysophospholipid transport from the lysosome.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8ZSU
 
 | | Lysozyme Crystallized with Bioassembler in Space Microgravity | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | MacCarthy, C, Koudan, E.V, Petrov, S.V, Levin, A.A, Khesuani, Y.D, Borshchevskiy, V. | | Deposit date: | 2024-06-05 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Exploring the potential of a bioassembler for protein crystallization in space.
Npj Microgravity, 11, 2025
|
|
7G03
 
 | | Crystal Structure of human FABP4 in complex with 6-fluoro-1,3-benzothiazol-2-amine | | Descriptor: | 6-fluoro-1,3-benzothiazol-2-amine, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Winternitz, P, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
8ZST
 
 | | Lysozyme Crystallized with Bioassembler on Earth | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | MacCarthy, C, Koudan, E.V, Petrov, S.V, Levin, A.A, Khesuani, Y.D, Borshchevskiy, V. | | Deposit date: | 2024-06-05 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Exploring the potential of a bioassembler for protein crystallization in space.
Npj Microgravity, 11, 2025
|
|
7FX4
 
 | | Crystal Structure of human FABP4 in complex with rac-(1R,2S)-2-[(3,4-dichlorophenoxy)methyl]cyclohexane-1-carboxylic acid, i.e. SMILES C1CC[C@H]([C@H](C1)C(=O)O)COc1cc(c(cc1)Cl)Cl with IC50=3.73482 microM | | Descriptor: | (1R,2S)-2-[(3,4-dichlorophenoxy)methyl]cyclohexane-1-carboxylic acid, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Buettelmann, B, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7P63
 
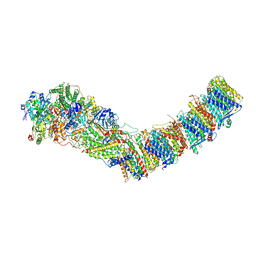 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 6, Closed state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P69
 
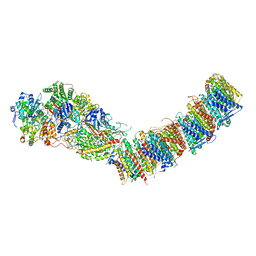 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 6, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7FXR
 
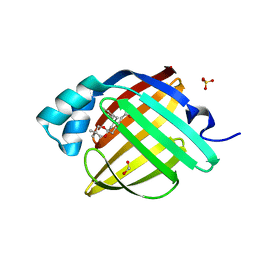 | | Crystal Structure of human FABP4 in complex with 2-[rac-(9R,10S)-10-benzyl-3,3-dimethyl-1,5-dioxaspiro[5.5]undecan-9-yl]acetic acid, i.e. SMILES C12(C[C@H]([C@@H](CC1)CC(=O)O)Cc1ccccc1)OCC(CO2)(C)C with IC50=0.106 microM | | Descriptor: | FORMIC ACID, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Chen, J, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7P7L
 
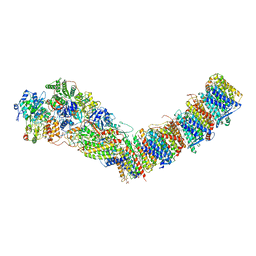 | | Complex I from E. coli, DDM/LMNG-purified, with NADH and FMN, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
6ULJ
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-phenyl-N-{6-[4-({6-[(phenylacetyl)amino]pyridazin-3-yl}oxy)piperidin-1-yl]pyridazin-3-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-08 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00045
To Be Published
|
|
7FWN
 
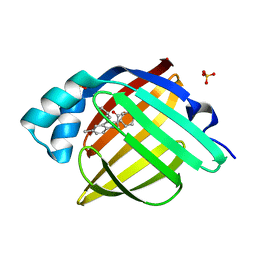 | | Crystal Structure of human FABP4 in complex with 3-(5-methyl-2,3-diphenylindol-1-yl)propanoic acid, i.e. SMILES N1(C(=C(c2c1ccc(c2)C)c1ccccc1)c1ccccc1)CCC(=O)O with IC50=0.038 microM | | Descriptor: | 3-(5-methyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5LZ8
 
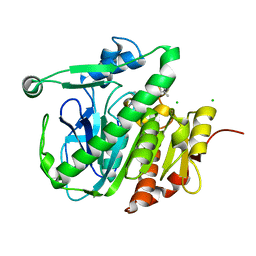 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | Descriptor: | 5-[2-[(4~{S})-4-~{tert}-butyl-2-oxidanylidene-pyrrolidin-1-yl]ethoxy]-2-fluoranyl-benzenecarbonitrile, CHLORIDE ION, Platelet-activating factor acetylhydrolase | | Authors: | Woolford, A, Day, P. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
7P61
 
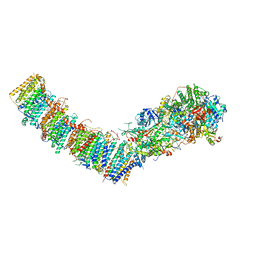 | | Complex I from E. coli, DDM-purified, with NADH, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P64
 
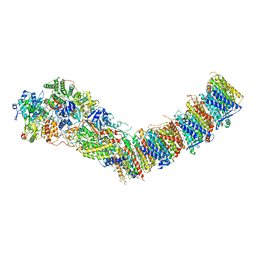 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 6, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7C
 
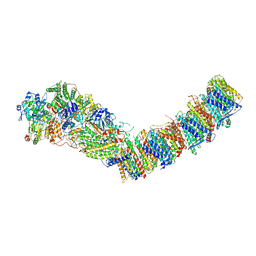 | | Complex I from E. coli, DDM/LMNG-purified, Apo, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7J
 
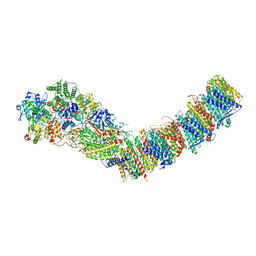 | | Complex I from E. coli, DDM/LMNG-purified, with DQ, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
5CYU
 
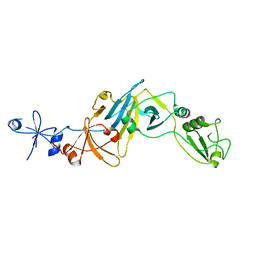 | | Structure of the soluble domain of EccB1 from the Mycobacterium smegmatis ESX-1 secretion system. | | Descriptor: | Conserved membrane protein | | Authors: | Arbing, M.A, Chan, S, Kahng, S, Kim, J, Eisenberg, D.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structures of EccB1 and EccD1 from the core complex of the mycobacterial ESX-1 type VII secretion system.
Bmc Struct.Biol., 16, 2016
|
|
7FYB
 
 | | Crystal Structure of human FABP4 in complex with rac-(1R,2S,4R)-2-[(3-chlorobenzoyl)amino]-4-phenoxycyclohexane-1-carboxylic acid, i.e. SMILES C1[C@H](C[C@@H]([C@@H](C1)C(=O)O)NC(=O)c1cc(ccc1)Cl)Oc1ccccc1 with IC50=4.26773 microM | | Descriptor: | (1R,2S,4R)-2-(3-chlorobenzamido)-4-phenoxycyclohexane-1-carboxylic acid, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Buettelmann, B, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FX7
 
 | | Crystal Structure of human FABP4 with binding site mutated to FABP5 in complex with 5-methylsulfanyl-2-[(4-phenylmethoxy-1H-indazol-3-yl)methylsulfanyl]pyrimidin-4-ol, i.e. SMILES n1c(c(cnc1SCC1=NNc2c1c(OCc1ccccc1)ccc2)SC)O with IC50=0.372 microM | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(benzyloxy)-1H-indazol-3-yl]methyl}sulfanyl)-5-(methylsulfanyl)pyrimidin-4-ol, DIMETHYL SULFOXIDE, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Lubbers, T, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5GQK
 
 | | Crystal structure of Cypovirus Polyhedra mutant with deletion of Gly192-Ala194 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Polyhedrin | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
5GQN
 
 | | Crystal structure of in cellulo Cypovirus Polyhedra mutant with deletion of Gly192-Ala194 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Polyhedrin | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
9GQ3
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(1,4)-(E) | | Descriptor: | (2~{S},9~{S},13~{E})-2-cyclohexyl-22,25-dimethoxy-11,16,20-trioxa-4-azatricyclo[19.2.2.0^{4,9}]pentacosa-1(24),13,21(25),22-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
6ILB
 
 | | Native crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi | | Descriptor: | 1,2-ETHANEDIOL, Fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi in complex with 1 glycerol, ZINC ION | | Authors: | Choi, M.Y, Kang, L.W, Ho, T.H, Nguyen, D.Q, Lee, I.H, Lee, J.H, Park, Y.S, Park, H.J. | | Deposit date: | 2018-10-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.50541973 Å) | | Cite: | Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi
To be published
|
|
5LYY
 
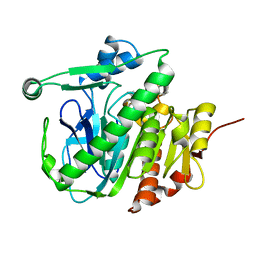 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | Descriptor: | 3-[2-(4-fluoranylphenoxy)ethyl]-1,3-diazaspiro[4.5]decane-2,4-dione, Platelet-activating factor acetylhydrolase | | Authors: | Woolford, A, Day, P. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
