5SP2
 
 | |
5SP3
 
 | |
5SP0
 
 | |
5SOL
 
 | |
4RRV
 
 | |
5SP9
 
 | |
5SP1
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC001472868186 | | Descriptor: | 3-{[methyl(pyrido[2,3-b]pyrazin-6-yl)amino]methyl}[1,2,4]triazolo[4,3-a]pyrazin-8(7H)-one, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SP7
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010903509 - (S,S) isomer | | Descriptor: | (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-1,2,3,4-tetrahydronaphthalene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SP6
 
 | |
5SP4
 
 | |
5SPC
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894387 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-4-hydroxy-1-[(5,6,7,8-tetrahydro-1,8-naphthyridine-3-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-[(5,6,7,8-tetrahydro-1,8-naphthyridine-3-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SP8
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894415 - (S) isomer | | Descriptor: | (6~{S})-7-[4-(cyclopropylcarbamoylamino)phenyl]carbonyl-3-methyl-6,8-dihydro-5~{H}-[1,2,4]triazolo[4,3-a]pyrazine-6-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SPD
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z4718398539 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-1-[4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SPE
 
 | |
5SPF
 
 | |
5SPG
 
 | |
5SPI
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z4574659604 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-1-{4-[(cyclopropanecarbonyl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-{4-[(cyclopropanecarbonyl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SPH
 
 | |
5SPK
 
 | |
5SPL
 
 | |
5CIE
 
 | |
4OOT
 
 | |
5CIF
 
 | |
1G4K
 
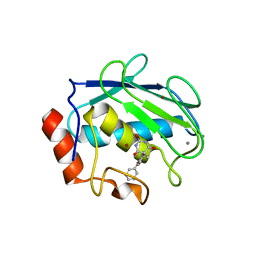 | | X-ray Structure of a Novel Matrix Metalloproteinase Inhibitor Complexed to Stromelysin | | Descriptor: | 5-METHYL-5-(4-PHENOXY-PHENYL)-PYRIMIDINE-2,4,6-TRIONE, CALCIUM ION, GLYCEROL, ... | | Authors: | Dunten, P, Kammlott, U, Crowther, R, Levin, W, Foley, L.H, Wang, P, Palermo, R. | | Deposit date: | 2000-10-27 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of a novel matrix metalloproteinase inhibitor complexed to stromelysin.
Protein Sci., 10, 2001
|
|
6BC8
 
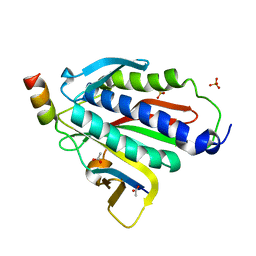 | | Crystal structure of Rev7-R124A/Rev3-RBM2 (residues 1988-2014) complex | | Descriptor: | ACETATE ION, DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B, ... | | Authors: | Rizzo, A.A, Hao, B, Li, Y, Korzhnev, D.M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
