7GTB
 
 | |
7GTJ
 
 | |
1H9J
 
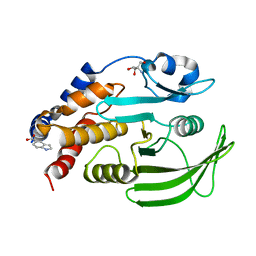
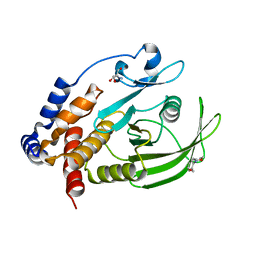
 | | Two crystal structures of the cytoplasmic molybdate-binding protein ModG suggest a novel cooperative binding mechanism and provide insights into ligand-binding specificity. Phosphate-grown form with molybdate and phosphate bound | | Descriptor: | MOLYBDATE ION, MOLYBDENUM-BINDING-PROTEIN, PHOSPHATE ION | | Authors: | Delarbre, L, Stevenson, C.E.M, White, D.J, Mitchenall, L.A, Pau, R.N, Lawson, D.M. | | Deposit date: | 2001-03-13 | | Release date: | 2001-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two Crystal Structures of the Cytoplasmic Molybdate-Binding Protein Modg Suggest a Novel Cooperative Binding Mechanism and Provide Insights Into Ligand-Binding Specificity
J.Mol.Biol., 308, 2001
|
|
7GTV
 
 | |
7GUC
 
 | |
7GSV
 
 | |
7GT3
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000527a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, ethyl (3R,3aS,8bS)-1-acetyl-5-methyl-2,3,3a,8b-tetrahydro-1H-[1]benzofuro[3,2-b]pyrrole-3-carboxylate | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GT1
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOMB000209a | | Descriptor: | (2S)-2-(2-chloro-6-fluorophenyl)-2,3-dihydroquinazolin-4(1H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
1HE1
 
 | | Crystal structure of the complex between the GAP domain of the Pseudomonas aeruginosa ExoS toxin and human Rac | | Descriptor: | ALUMINUM FLUORIDE, EXOENZYME S, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wurtele, M, Wolf, E, Pederson, K.J, Buchwald, G, Ahmadian, M.R, Barbieri, J.T, Wittinghofer, A. | | Deposit date: | 2000-11-18 | | Release date: | 2001-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How the Pseudomonas Aeruginosa Exos Toxin Downregulates Rac
Nat.Struct.Biol., 8, 2001
|
|
7GT6
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000530a | | Descriptor: | (3aS,8aS)-6-benzoyloctahydropyrrolo[3,4-d]azepin-1(2H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GT9
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMSOA000463b | | Descriptor: | (3R)-4-oxo-3,4-dihydro-2H-1-benzopyran-3-carbonitrile, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTI
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000571a | | Descriptor: | 1-{(1S,4R,5S,6R)-6-hydroxy-4-[(pyridin-2-yl)oxy]-2-azabicyclo[3.3.1]nonan-2-yl}ethan-1-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GSH
 
 | |
7GTE
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with XST00000646b | | Descriptor: | (5S)-5-(trifluoromethyl)-1,4-diazepane, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTM
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000543a | | Descriptor: | (4S)-4-hydroxy-2-(propan-2-yl)-3,4-dihydro-1lambda~6~,2-benzothiazine-1,1(2H)-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
1HGY
 
 | | CEL6A D221A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), alpha-D-glucopyranose, ... | | Authors: | Zou, J.-Y, Kleywegt, G.J, Jones, T.A. | | Deposit date: | 2000-12-15 | | Release date: | 2002-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site of Cellobiohydrolase Cel6A from Trichoderma Reesei: The Roles of Aspartic Acids D221 and D175
J.Am.Chem.Soc., 124, 2002
|
|
7GS8
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000466a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, ~{N},~{N},5,6-tetramethylthieno[2,3-d]pyrimidin-4-amine | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GSJ
 
 | |
7GSX
 
 | |
7GT8
 
 | |
1H88
 
 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX1 | | Descriptor: | AMMONIUM ION, CCAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-29 | | Release date: | 2002-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
7GSI
 
 | |
7GTC
 
 | |
1H0I
 
 | | Complex of a chitinase with the natural product cyclopentapeptide argifin from Gliocladium | | Descriptor: | ARGIFIN, CHITINASE B, GLYCEROL, ... | | Authors: | Houston, D.R, Shiomi, K, Arai, N, Omura, S, Peter, M.G, Turberg, A, Synstad, B, Eijsink, V.G.H, Aalten, D.M.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Inhibited Complexes of a Chitinase with Natural Product Cyclopentapeptides - Peptide Mimicry of a Carbohydrate Substrate
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7GTY
 
 | |
