7ZBR
 
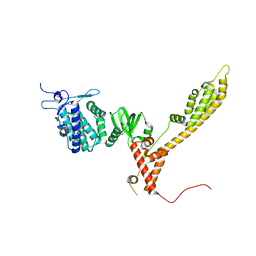 | | Crystal Structure of full-length Bartonella Effector protein 1 | | Descriptor: | Bartonella effector protein (Bep) substrate of VirB T4SS | | Authors: | Huber, M. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Full-length structure of a FIC effector protein from Bartonella reveals a novel structural domain
To be published
|
|
7L64
 
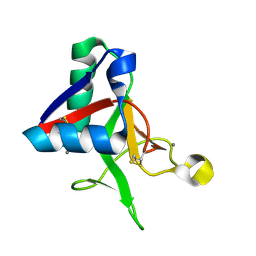 | |
7PVP
 
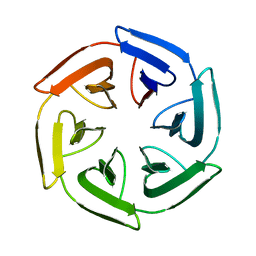 | | Crystal structure of the SAKe6BC designer protein | | Descriptor: | SAKe6BC | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-10-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
8RTE
 
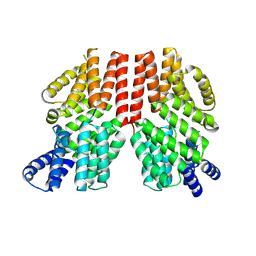 | |
7A9B
 
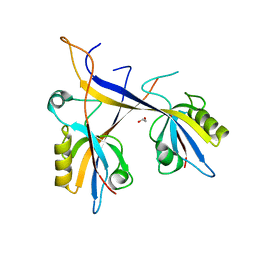 | | Crystal structure of Shank1 PDZ domain with ARAP3-derived peptide | | Descriptor: | 1,2-ETHANEDIOL, SH3 and multiple ankyrin repeat domains protein 1,Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Mariam McAuley, M, Ali, M, Ivarsson, Y, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-01 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Shank1 PDZ domain with ARAP3-derived peptide
to be published
|
|
8RJ4
 
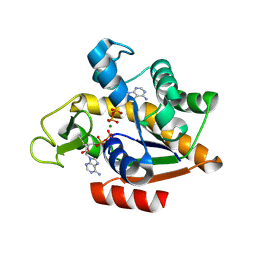 | | E. coli adenylate kinase in complex with two ADP molecules and Mg2+ as a result of enzymatic AP4A hydrolysis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase, CHLORIDE ION, ... | | Authors: | Tischlik, S, Ronge, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2023-12-20 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Magnesium induced structural reorganization in the active site of adenylate kinase.
Sci Adv, 10, 2024
|
|
7LDM
 
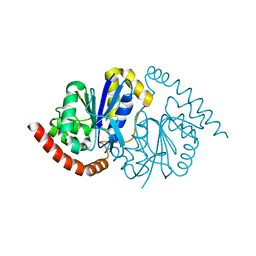 | | G150T Pseudomonas fluorescens isocyanide hydratase (G150T-1) at 274K, Phenix-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7Z82
 
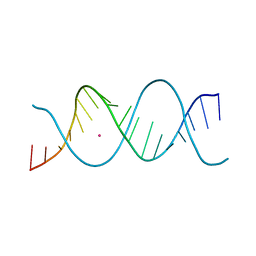 | | REP-related Chom18 variant with double AG mismatch | | Descriptor: | Chom18-AG DNA, STRONTIUM ION | | Authors: | Svoboda, J, Kolenko, P, Berdar, D, Schneider, B. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8RD1
 
 | | HUWE1 WWE domain in complex with compound 4 | | Descriptor: | 2-(2-hydroxy-2-oxoethyl)-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid, CHLORIDE ION, E3 ubiquitin-protein ligase HUWE1 | | Authors: | Muenzker, L, Zak, K.M, Boettcher, J. | | Deposit date: | 2023-12-07 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Commun Biol, 7, 2024
|
|
7LF5
 
 | | Structure of Hyperglycosylated Human IgG1 Fc (Fc267) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgG1 Fc (Fc267), alpha-L-fucopyranose, ... | | Authors: | Fields, J.K, Sundberg, E.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Silent Antibodies: Generation of Hyperglycosylated FCs to Ablate Effector Functions
To Be Published
|
|
6HZG
 
 | | BP0997, GH138 enzyme targeting pectin rhamnogalacturonan II | | Descriptor: | BPa0997 N-ter E361S, CHLORIDE ION, SODIUM ION, ... | | Authors: | Basle, A, Cartmell, A, Labourel, A, Gilbert, H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analyses of glycoside hydrolase 138 enzymes targeting chain A galacturonic acid in the complex pectin rhamnogalacturonan II.
J.Biol.Chem., 294, 2019
|
|
7L61
 
 | |
8RG8
 
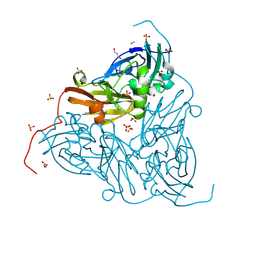 | | High pH (8.0) nitrite-bound MSOX movie series dataset 1 of the copper nitrite reductase from Bradyrhizobium sp. ORS375 (two-domain) [0.68 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
7ZA0
 
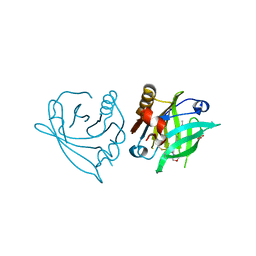 | |
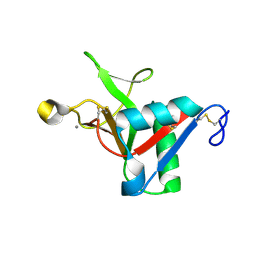
7L67
 
 | |
6I05
 
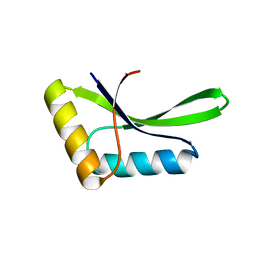 | |
8RU9
 
 | | High pH (8.0) nitrite-bound MSOX movie series dataset 1 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [0.69 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-01-30 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
7LFP
 
 | | SARS-CoV-2 Main protease immature form - F2X Entry Library G05 fragment | | Descriptor: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-01-18 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
8RFO
 
 | | Low pH (pH 5.5) nitrite-bound MSOX movie series dataset 2 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [1.22 MGy] - nitrite intermediate | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
7Z7Z
 
 | | REP-related Chom18 variant with double TA base pair | | Descriptor: | Chom18-TA DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7LBH
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-1) at 274K, PHENIX-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-08 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7ADR
 
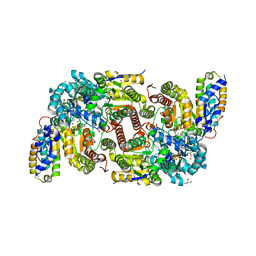 | | CO bound as bridging ligand at the active site of vanadium nitrogenase VFe protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Rohde, M, Grunau, K, Einsle, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | CO Binding to the FeV Cofactor of CO-Reducing Vanadium Nitrogenase at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6BHG
 
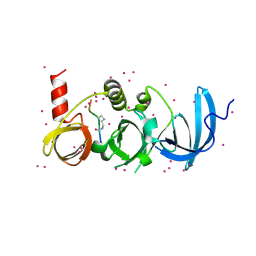 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
7LBL
 
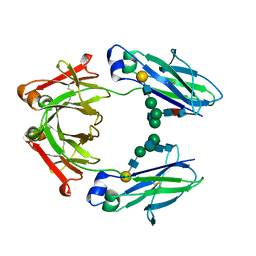 | | Structure of Human IgG1 Fc | | Descriptor: | Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Fields, J.K, Sundberg, E.J. | | Deposit date: | 2021-01-08 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Silent Antibodies: Generation of Hyperglycosylated FCs to Ablate Effector Functions
To Be Published
|
|
6I0Q
 
 | |
