6ZIQ
 
 | | bovine ATP synthase stator domain, state 1 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C1, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (4.33 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZIT
 
 | | bovine ATP synthase Stator domain, state 2 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C2, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZPO
 
 | | bovine ATP synthase monomer state 1 (combined) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZMR
 
 | | Porcine ATP synthase Fo domain | | Descriptor: | ATP synthase F(0) complex subunit C1, mitochondrial, ATP synthase g subunit, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-07-03 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZQN
 
 | | bovine ATP synthase monomer state 3 (combined) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-07-10 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8DBR
 
 | | E. coli ATP synthase imaged in 10mM MgATP State2 "half-up | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
8DBT
 
 | | E. coli ATP synthase imaged in 10mM MgATP State2 "down | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
8DBP
 
 | | E. coli ATP synthase imaged in 10mM MgATP State1 "half-up | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
8DBS
 
 | |
8DBW
 
 | |
8DBU
 
 | |
8DBQ
 
 | |
8DBV
 
 | | E. coli ATP synthase imaged in 10mM MgATP State3 "down | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
8F29
 
 | | Yeast ATP synthase in conformation-1 at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-07 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8F39
 
 | | Yeast ATP synthase in conformation-2, at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8G08
 
 | |
8G09
 
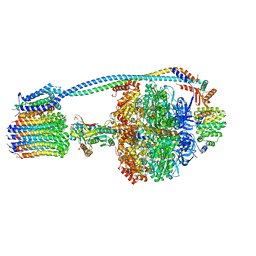 | |
8G0B
 
 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase FO region | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ATP synthase subunit a, ATP synthase subunit b, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8G0C
 
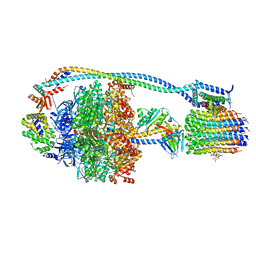 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 1 (backbone model) | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8G0E
 
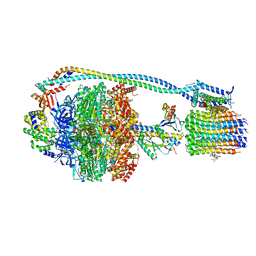 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 3 | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8G0A
 
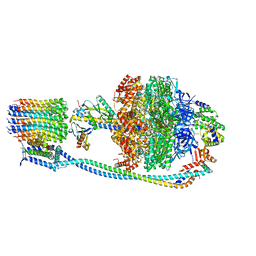 | |
8G07
 
 | |
8G0D
 
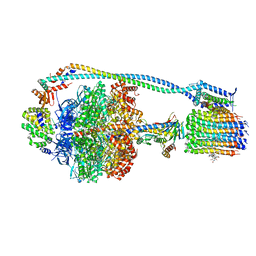 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
6O7V
 
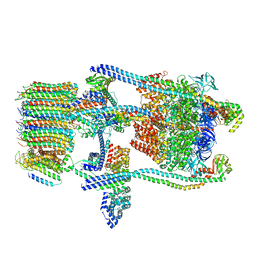 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 1 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6OQW
 
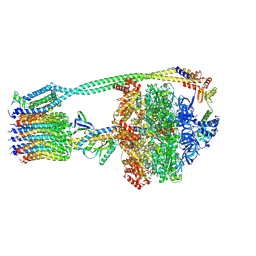 | | E. coli ATP synthase State 3a | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
