3L4P
 
 | |
3LB8
 
 | |
1N60
 
 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Cyanide-inactivated Form | | Descriptor: | Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, Carbon monoxide dehydrogenase small chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N5X
 
 | | Xanthine Dehydrogenase from Bovine Milk with Inhibitor TEI-6720 Bound | | Descriptor: | 2-(3-CYANO-4-ISOBUTOXY-PHENYL)-4-METHYL-5-THIAZOLE-CARBOXYLIC ACID, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Okamoto, K, Eger, B.T, Nishino, T, Kondo, S, Pai, E.F, Nishino, T. | | Deposit date: | 2002-11-07 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An Extremely Potent Inhibitor of Xanthine Oxidoreductase: Crystal Structure of the Enzyme-Inhibitor Complex and Mechanism of Inhibition
J.BIOL.CHEM., 278, 2003
|
|
1N62
 
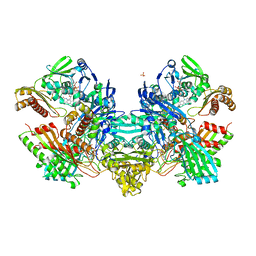 | | Crystal Structure of the Mo,Cu-CO Dehydrogenase (CODH), n-butylisocyanide-bound state | | Descriptor: | CU(I)-S-MO(IV)(=O)O-NBIC CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JRP
 
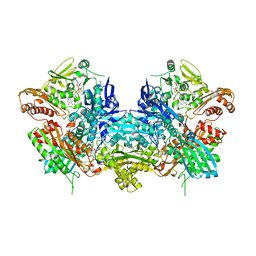 | | Crystal Structure of Xanthine Dehydrogenase inhibited by alloxanthine from Rhodobacter capsulatus | | Descriptor: | CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Truglio, J.J, Theis, K, Leimkuhler, S, Rappa, R, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2001-08-14 | | Release date: | 2002-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the active and alloxanthine-inhibited forms of xanthine dehydrogenase from Rhodobacter capsulatus
Structure, 10, 2002
|
|
1JRO
 
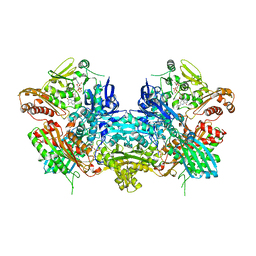 | | Crystal Structure of Xanthine Dehydrogenase from Rhodobacter capsulatus | | Descriptor: | CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Truglio, J.J, Theis, K, Leimkuhler, S, Rappa, R, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2001-08-14 | | Release date: | 2002-01-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the active and alloxanthine-inhibited forms of xanthine dehydrogenase from Rhodobacter capsulatus
Structure, 10, 2002
|
|
1N5W
 
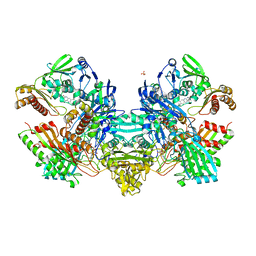 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Oxidized form | | Descriptor: | CU(I)-S-MO(VI)(=O)OH CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-07 | | Release date: | 2002-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N63
 
 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Carbon monoxide reduced state | | Descriptor: | CU(I)-S-MO(IV)(=O)OH CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N61
 
 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Dithionite reduced state | | Descriptor: | CU(I)-S-MO(IV)(=O)OH CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3NVZ
 
 | | Crystal Structure of Bovine Xanthine Oxidase in Complex with Indole-3-Aldehyde | | Descriptor: | 1H-INDOLE-3-CARBALDEHYDE, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Cao, H, Hille, R. | | Deposit date: | 2010-07-08 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate orientation and specificity in xanthine oxidase: crystal structures of the enzyme in complex with indole-3-acetaldehyde and guanine.
Biochemistry, 53, 2014
|
|
3NVY
 
 | | Crystal Structure of Bovine Xanthine Oxidase in Complex with Quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Cao, H, Hille, R. | | Deposit date: | 2010-07-08 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray Crystal Structure of a Xanthine Oxidase Complex with the Flavonoid Inhibitor Quercetin.
J Nat Prod, 77, 2014
|
|
3NRZ
 
 | | Crystal Structure of Bovine Xanthine Oxidase in Complex with Hypoxanthine | | Descriptor: | DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cao, H, Pauff, J.M, Hille, R. | | Deposit date: | 2010-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate orientation and catalytic specificity in the action of xanthine oxidase: the sequential hydroxylation of hypoxanthine to uric acid.
J.Biol.Chem., 285, 2010
|
|
1KRH
 
 | | X-ray Structure of Benzoate Dioxygenase Reductase | | Descriptor: | Benzoate 1,2-Dioxygenase Reductase, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Karlsson, A, Beharry, Z.M, Eby, D.M, Coulter, E.D, Niedle, E.L, Kurtz Jr, D.M, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-01-09 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of benzoate 1,2-dioxygenase reductase from Acinetobacter sp. strain ADP1.
J.Mol.Biol., 318, 2002
|
|
9LRR
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase NqrB-G141A mutant from Vibrio cholerae with bound korormicin A | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-02-01 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural Elucidation of the Mechanism for Inhibitor Resistance in the Na + -Translocating NADH-Ubiquinone Oxidoreductase from Vibrio cholerae.
Biochemistry, 64, 2025
|
|
5Y6Q
 
 | |
9U5G
 
 | |
9UD2
 
 | |
9UD3
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase NqrB-T236Y mutant from Vibrio cholerae | | Descriptor: | CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-06 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9UD4
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase NqrB-T236Y mutant from Vibrio cholerae reduced by NADH | | Descriptor: | CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-06 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9UD9
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase from Vibrio cholerae reduced by NADH, in the absence of Na+, down state | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-07 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9UD6
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase from Vibrio cholerae reduced by NADH, in the absence of Na+, upper state | | Descriptor: | CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-06 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9UDA
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase NqrB-G141A mutant from Vibrio cholerae reduced by NADH, with bound korormicin A, stable state | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-06 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9UD5
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase from Vibrio cholerae reduced by NADH, with bound korormicin A | | Descriptor: | CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-06 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9UDF
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase NqrB-G141A mutant from Vibrio cholerae reduced by NADH, with bound korormicin A, shifted state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-06 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
