1SRR
 
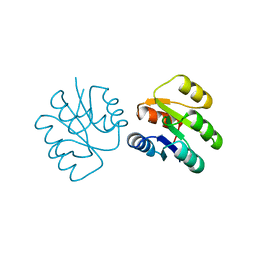 | | CRYSTAL STRUCTURE OF A PHOSPHATASE RESISTANT MUTANT OF SPORULATION RESPONSE REGULATOR SPO0F FROM BACILLUS SUBTILIS | | Descriptor: | CALCIUM ION, SPORULATION RESPONSE REGULATORY PROTEIN | | Authors: | Madhusudan, Whiteley, J.M, Hoch, J.A, Zapf, J, Xuong, N.H, Varughese, K.I. | | Deposit date: | 1996-04-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a phosphatase-resistant mutant of sporulation response regulator Spo0F from Bacillus subtilis.
Structure, 4, 1996
|
|
1SRS
 
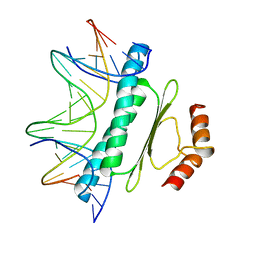 | | SERUM RESPONSE FACTOR (SRF) CORE COMPLEXED WITH SPECIFIC SRE DNA | | Descriptor: | DNA (5'-D(*CP*CP*(5IU)P*TP*CP*CP*TP*AP*AP*TP*TP*AP*GP*GP*CP*CP*AP*TP*G)-3'), DNA (5'-D(*CP*CP*AP*TP*GP*GP*CP*CP*TP*AP*AP*TP*TP*AP*GP*GP*A P*AP*G)-3'), PROTEIN (SERUM RESPONSE FACTOR (SRF)) | | Authors: | Pellegrini, L, Tan, S, Richmond, T.J. | | Deposit date: | 1995-07-28 | | Release date: | 1995-07-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of serum response factor core bound to DNA.
Nature, 376, 1995
|
|
1SRU
 
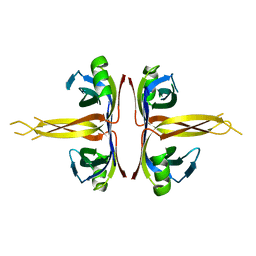 | | Crystal structure of full length E. coli SSB protein | | Descriptor: | Single-strand binding protein | | Authors: | Savvides, S.N, Raghunathan, S, Fuetterer, K, Kozlov, A.G, Lohman, T.M, Waksman, G. | | Deposit date: | 2004-03-23 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA.
Protein Sci., 13, 2004
|
|
1SRV
 
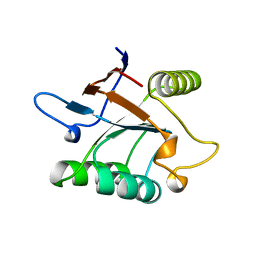 | | THERMUS THERMOPHILUS GROEL (HSP60 CLASS) FRAGMENT (APICAL DOMAIN) COMPRISING RESIDUES 192-336 | | Descriptor: | PROTEIN (GROEL (HSP60 CLASS)) | | Authors: | Walsh, M.A, Dementieva, I, Evans, G, Sanishvili, R, Joachimiak, A. | | Deposit date: | 1999-03-02 | | Release date: | 1999-03-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Taking MAD to the extreme: ultrafast protein structure determination.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1SRX
 
 | |
1SRY
 
 | |
1SRZ
 
 | | Solution structure of the second complement control protein (CCP) module of the GABA(B)R1a receptor, Pro-119 trans conformer | | Descriptor: | Gamma-aminobutyric acid type B receptor, subunit 1 | | Authors: | Blein, S, Uhrin, D, Smith, B.O, White, J.H, Barlow, P.N. | | Deposit date: | 2004-03-23 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Complement Control Protein (CCP) Modules of GABAB Receptor 1a: ONLY ONE OF THE TWO CCP MODULES IS COMPACTLY FOLDED
J.Biol.Chem., 279, 2004
|
|
1SS1
 
 | | STAPHYLOCOCCAL PROTEIN A, B-DOMAIN, Y15W MUTANT, NMR, 25 STRUCTURES | | Descriptor: | Immunoglobulin G binding protein A | | Authors: | Sato, S, Religa, T.L, Daggett, V, Fersht, A.R. | | Deposit date: | 2004-03-23 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From The Cover: Testing protein-folding simulations by experiment: B domain of protein A.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SS2
 
 | | Solution structure of the second complement control protein (CCP) module of the GABA(B)R1a receptor, Pro-119 cis conformer | | Descriptor: | Gamma-aminobutyric acid type B receptor, subunit 1 | | Authors: | Blein, S, Uhrin, D, Smith, B.O, White, J.H, Barlow, P.N. | | Deposit date: | 2004-03-23 | | Release date: | 2004-10-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the complement control protein (CCP) modules of GABA(B) receptor 1a: only one of the two CCP modules is compactly folded.
J.Biol.Chem., 279, 2004
|
|
1SS3
 
 | | Solution structure of Ole e 6, an allergen from olive tree pollen | | Descriptor: | Pollen allergen Ole e 6 | | Authors: | Trevino, M.A, Garcia-Mayoral, M.F, Barral, P, Villalba, M, Santoro, J, Rico, M, Rodriguez, R, Bruix, M. | | Deposit date: | 2004-03-23 | | Release date: | 2004-08-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Ole e 6, a Major Allergen from Olive Tree Pollen.
J.Biol.Chem., 279, 2004
|
|
1SS4
 
 | |
1SS6
 
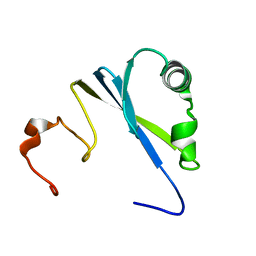 | | Solution structure of SEP domain from human p47 | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Soukenik, M, Leidert, M, Sievert, V, Buessow, K, Leitner, D, Labudde, D, Ball, L.J, Oschkinat, H. | | Deposit date: | 2004-03-23 | | Release date: | 2004-11-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The SEP domain of p47 acts as a reversible competitive inhibitor of cathepsin L
FEBS Lett., 576, 2004
|
|
1SS7
 
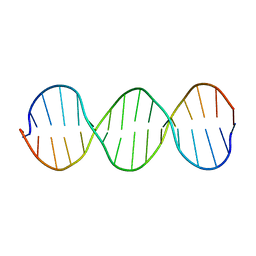 | | Compensating bends in a 16 base-pair DNA oligomer containing a T3A3 segment | | Descriptor: | 5'-D(*CP*GP*AP*GP*GP*TP*TP*TP*AP*AP*AP*CP*CP*TP*CP*G)-3' | | Authors: | McAteer, K, Aceves-Gaona, A, Michalczyk, R, Buchko, G.W, Isern, N.G, Silks, L.A, Miller, J.H, Kennedy, M.A. | | Deposit date: | 2004-03-23 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Compensating bends in a 16-base-pair DNA oligomer containing a T(3)A(3) segment: A NMR study of global DNA curvature
Biopolymers, 75, 2004
|
|
1SS8
 
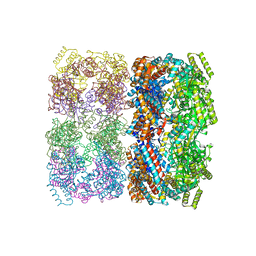 | | GroEL | | Descriptor: | groEL protein | | Authors: | Chaudhry, C, Horwich, A.L, Brunger, A.T, Adams, P.D. | | Deposit date: | 2004-03-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Exploring the structural dynamics of the E.coli chaperonin GroEL using translation-libration-screw crystallographic refinement of intermediate states.
J.Mol.Biol., 342, 2004
|
|
1SS9
 
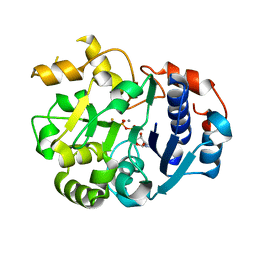 | | Crystal Structural Analysis of Active Site Mutant Q189E of LgtC | | Descriptor: | MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUOROGALACTOSE, alpha-1,4-galactosyl transferase | | Authors: | Lairson, L.L, Chiu, C.P, Ly, H.D, He, S, Wakarchuk, W.W, Strynadka, N.C, Withers, S.G. | | Deposit date: | 2004-03-23 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Intermediate trapping on a mutant retaining alpha-galactosyltransferase identifies an unexpected aspartate residue.
J.Biol.Chem., 279, 2004
|
|
1SSA
 
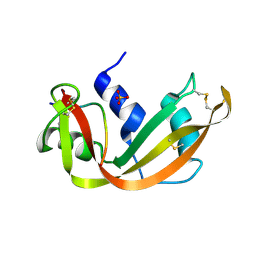 | | A STRUCTURAL INVESTIGATION OF CATALYTICALLY MODIFIED F12OL AND F12OY SEMISYNTHETIC RIBONUCLEASES | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | deMel, V.S.J, Doscher, M.S, Glinn, M.A, Martin, P.D, Ram, M.L, Edwards, B.F.P. | | Deposit date: | 1993-08-03 | | Release date: | 1994-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of catalytically modified F120L and F120Y semisynthetic ribonucleases.
Protein Sci., 3, 1994
|
|
1SSB
 
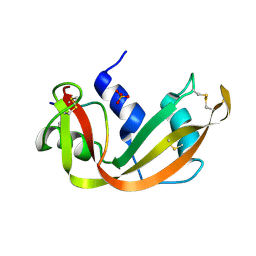 | | A STRUCTURAL INVESTIGATION OF CATALYTICALLY MODIFIED F12OL AND F12OY SEMISYNTHETIC RIBONUCLEASES | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Demel, V.S.J, Doscher, M.S, Glinn, M.A, Martin, P.D, Ram, M.L, Edwards, B.F.P. | | Deposit date: | 1993-08-03 | | Release date: | 1994-09-30 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of catalytically modified F120L and F120Y semisynthetic ribonucleases.
Protein Sci., 3, 1994
|
|
1SSC
 
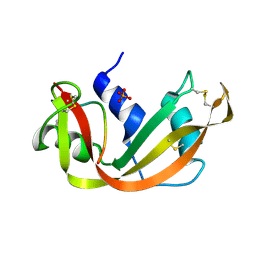 | | THE 1.6 ANGSTROMS STRUCTURE OF A SEMISYNTHETIC RIBONUCLEASE CRYSTALLIZED FROM AQUEOUS ETHANOL. COMPARISON WITH CRYSTALS FROM SALT SOLUTIONS AND WITH RNASE A FROM AQUEOUS ALCOHOL SOLUTIONS | | Descriptor: | PHOSPHATE ION, RIBONUCLEASE A | | Authors: | De Mel, V.S.J, Doscher, M.S, Martin, P.D, Rodier, F, Edwards, B.F.P. | | Deposit date: | 1994-10-05 | | Release date: | 1995-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1.6 A structure of semisynthetic ribonuclease crystallized from aqueous ethanol. Comparison with crystals from salt solutions and with ribonuclease A from aqueous alcohol solutions.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1SSD
 
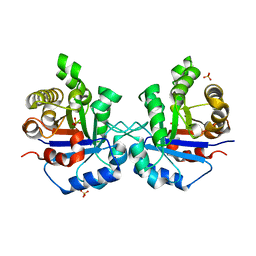 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | SULFATE ION, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SSE
 
 | |
1SSF
 
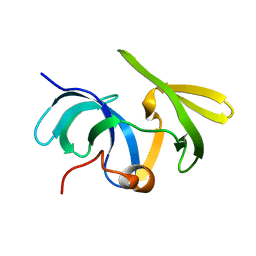 | | Solution structure of the mouse 53BP1 fragment (residues 1463-1617) | | Descriptor: | Transformation related protein 53 binding protein 1 | | Authors: | Charier, G, Couprie, J, Alpha-Bazin, B, Meyer, V, Quemeneur, E, Guerois, R, Callebaut, I, Gilquin, B, Zinn-Justin, S. | | Deposit date: | 2004-03-24 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Tudor Tandem of 53BP1; A New Structural Motif Involved in DNA and RG-Rich Peptide Binding
Structure, 12, 2004
|
|
1SSG
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SSH
 
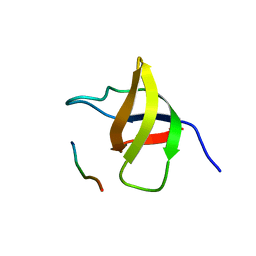 | | Crystal structure of the SH3 domain from a S. cerevisiae hypothetical 40.4 kDa protein in complex with a peptide | | Descriptor: | 12-mer peptide from Cytoskeleton assembly control protein SLA1, Hypothetical 40.4 kDa protein in PES4-HIS2 intergenic region | | Authors: | Kursula, P, Kursula, I, Lehmann, F, Song, Y.-H, Wilmanns, M. | | Deposit date: | 2004-03-24 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Yeast SH3 domain structural genomics
To be Published
|
|
1SSJ
 
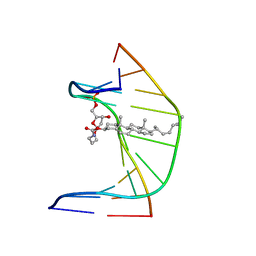 | | A DNA DUPLEX CONTAINING A CHOLESTEROL ADDUCT (BETA-ANOMER) | | Descriptor: | 5'-D(*CP*CP*AP*CP*(HOB)P*GP*GP*AP*AP*C)-3', 5'-D(GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3' | | Authors: | Gomez-Pinto, I, Cubero, E, Kalko, S.G, Monaco, V, van der Marel, G, van Boom, J.H, Orozco, M, Gonzalez, C. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Effect of bulky lesions on DNA: Solution structure of a DNA duplex containing a cholesterol adduct.
J.Biol.Chem., 279, 2004
|
|
1SSK
 
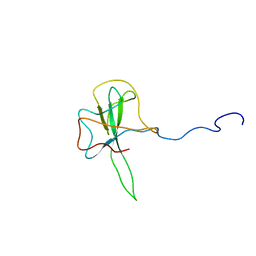 | | Structure of the N-terminal RNA-binding Domain of the SARS CoV Nucleocapsid Protein | | Descriptor: | Nucleocapsid protein | | Authors: | Huang, Q, Yu, L, Petros, A.M, Gunasekera, A, Liu, Z, Xu, N, Hajduk, P, Mack, J, Fesik, S.W, Olejniczak, E.T. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-Terminal RNA-Binding Domain of the SARS CoV Nucleocapsid Protein.
Biochemistry, 43, 2004
|
|
