1JBN
 
 | | Solution structure of an acyclic permutant of SFTI-1, A trypsin inhibitor from sunflower seeds | | Descriptor: | CYCLIC TRYPSIN INHIBITOR | | Authors: | Korsinczky, M.L.J, Schirra, H.J, Rosengren, K.J, West, J, Condie, B.A, Otvos, L, Anderson, M.A, Craik, D.J. | | Deposit date: | 2001-06-06 | | Release date: | 2001-08-22 | | Last modified: | 2016-12-28 | | Method: | SOLUTION NMR | | Cite: | Solution structures by 1H NMR of the novel cyclic trypsin inhibitor SFTI-1 from sunflower seeds and an acyclic permutant.
J.Mol.Biol., 311, 2001
|
|
1FTT
 
 | | THYROID TRANSCRIPTION FACTOR 1 HOMEODOMAIN (RATTUS NORVEGICUS) | | Descriptor: | THYROID TRANSCRIPTION FACTOR 1 HOMEODOMAIN | | Authors: | Fogolari, F, Esposito, G, Damante, G, Formisano, S, Di Lauro, R, Viglino, P. | | Deposit date: | 1995-10-03 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Analysis of the solution structure of the homeodomain of rat thyroid transcription factor 1 by 1H-NMR spectroscopy and restrained molecular mechanics.
Eur.J.Biochem., 241, 1996
|
|
5I1X
 
 | |
3KPX
 
 | | Crystal Structure Analysis of photoprotein clytin | | Descriptor: | Apophotoprotein clytin-3, C2-HYDROPEROXY-COELENTERAZINE, CALCIUM ION | | Authors: | Titushin, M.S, Li, Y, Stepanyuk, G.A, Wang, B.-C, Lee, J, Vysotski, E.S, Liu, Z.-J. | | Deposit date: | 2009-11-17 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | NMR derived topology of a GFP-photoprotein energy transfer complex
J.Biol.Chem., 285, 2010
|
|
1DCJ
 
 | | SOLUTION STRUCTURE OF YHHP, A NOVEL ESCHERICHIA COLI PROTEIN IMPLICATED IN THE CELL DIVISION | | Descriptor: | YHHP PROTEIN | | Authors: | Katoh, E, Hatta, T, Shindo, H, Mizuno, T, Yamazaki, T. | | Deposit date: | 1999-11-05 | | Release date: | 2001-07-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High precision NMR structure of YhhP, a novel Escherichia coli protein implicated in cell division.
J.Mol.Biol., 304, 2000
|
|
1DSV
 
 | | STRUCTURE OF THE MMTV NUCLEOCAPSID PROTEIN (C-TERMINAL ZINC FINGER) | | Descriptor: | NUCLEIC ACID BINDING PROTEIN P14, ZINC ION | | Authors: | Klein, D.J, Johnson, P.E, Zollars, E.S, De Guzman, R.N, Summers, M.F. | | Deposit date: | 2000-01-08 | | Release date: | 2000-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle.
Biochemistry, 39, 2000
|
|
1DSQ
 
 | | STRUCTURE OF THE MMTV NUCLEOCAPSID PROTEIN (ZINC FINGER 1) | | Descriptor: | NUCLEIC ACID BINDING PROTEIN P14, ZINC ION | | Authors: | Klein, D.J, Johnson, P.E, Zollars, E.S, De Guzman, R.N, Summers, M.F. | | Deposit date: | 2000-01-08 | | Release date: | 2000-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle.
Biochemistry, 39, 2000
|
|
1IY3
 
 | | Solution Structure of the Human lysozyme at 4 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
7MLA
 
 | | Solution NMR structure of HDMX in complex with Zn and MCo-52-2 | | Descriptor: | MCo-52-2, Protein Mdm4, ZINC ION | | Authors: | Ramirez, L.S, Theophall, G, Chaudhuri, D, Camarero, J.C, Shekhtman, A. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-04 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of HDMX in complex with Zn and cyclotide 52-2
To Be Published
|
|
1IY4
 
 | | Solution structure of the human lysozyme at 35 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
1DUF
 
 | | THE NMR STRUCTURE OF DNA DODECAMER DETERMINED IN AQUEOUS DILUTE LIQUID CRYSTALLINE PHASE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Tjandra, N, Tate, S, Ono, A, Kainosho, M, Bax, A. | | Deposit date: | 2000-01-17 | | Release date: | 2000-07-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of a DNA Dodecamer in an Aqueous Dilute Liquid Crystalline Phase
J.Am.Chem.Soc., 122, 2000
|
|
1BM4
 
 | | MOMLV CAPSID PROTEIN MAJOR HOMOLOGY REGION PEPTIDE ANALOG | | Descriptor: | PROTEIN (MOLONEY MURINE LEUKEMIA VIRUS CAPSID) | | Authors: | Clish, C.B, Peyton, D.H, Barklis, E. | | Deposit date: | 1998-07-28 | | Release date: | 1998-08-05 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human immunodeficiency virus type 1 (HIV-1) and moloney murine leukemia virus (MoMLV) capsid protein major-homology-region peptide analogs by NMR spectroscopy.
Eur.J.Biochem., 257, 1998
|
|
1IRP
 
 | |
1BN9
 
 | | RESPONSE ELEMENT OF THE ORPHAN NUCLEAR RECEPTOR REV-ERB BETA | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*CP*AP*TP*TP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*AP*AP*TP*GP*TP*AP*GP*GP*TP*CP*AP*G)-3') | | Authors: | Castagne, C, Terenzi, H, Zakin, M.M, Delepierre, M. | | Deposit date: | 1998-07-31 | | Release date: | 1998-08-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the orphan nuclear receptor rev-erb beta response element by 1H, 31P NMR and molecular simulation
Biochimie, 82, 2000
|
|
1DFS
 
 | | SOLUTION STRUCTURE OF THE ALPHA-DOMAIN OF MOUSE METALLOTHIONEIN-1 | | Descriptor: | CADMIUM ION, METALLOTHIONEIN-1 | | Authors: | Zangger, K, Oz, G, Otvos, J.D, Armitage, I.M. | | Deposit date: | 1999-11-20 | | Release date: | 1999-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mouse [Cd7]-metallothionein-1 by homonuclear and heteronuclear NMR spectroscopy.
Protein Sci., 8, 1999
|
|
1PQN
 
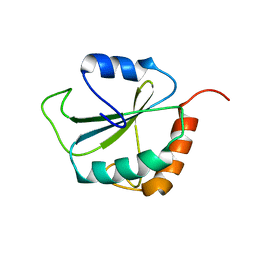 | | dominant negative human hDim1 (hDim1 1-128) | | Descriptor: | Spliceosomal U5 snRNP-specific 15 kDa protein | | Authors: | Zhang, Y.Z, Cheng, H, Gould, K.L, Golemis, E.A, Roder, H. | | Deposit date: | 2003-06-18 | | Release date: | 2003-08-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, stability and function of hDim1 investigated by NMR, circular
dichroism and mutational analysis
Biochemistry, 42, 2003
|
|
1IYM
 
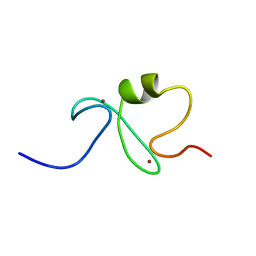 | | RING-H2 finger domain of EL5 | | Descriptor: | EL5, ZINC ION | | Authors: | Katoh, E, Katoh, S, Minami, E, Yamazaki, T. | | Deposit date: | 2002-08-30 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High Precision NMR Structure and Function of the RING-H2 Finger Domain of EL5, a Rice Protein Whose Expression Is Increased upon Exposure to Pathogen-derived Oligosaccharides
J.Biol.Chem., 278, 2003
|
|
1PFH
 
 | |
1G2G
 
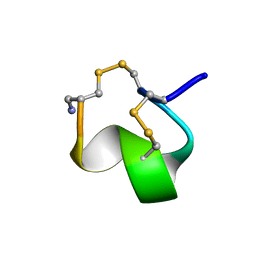 | | MINIMAL CONFORMATION OF THE ALPHA-CONOTOXIN IMI FOR THE ALPHA7 NEURONAL NICOTINIC ACETYLCHOLINE RECEPTOR RECOGNITION | | Descriptor: | ALPHA-CONOTOXIN IMI | | Authors: | Lamthanh, H, Jegou-Matheron, C, Servent, D, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-19 | | Release date: | 2000-11-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Minimal conformation of the alpha-conotoxin ImI for the alpha7 neuronal nicotinic acetylcholine receptor recognition: correlated CD, NMR and binding studies.
FEBS Lett., 454, 1999
|
|
6LEK
 
 | | Tertiary structure of Barnacle cement protein MrCP20 | | Descriptor: | Cement protein-20k | | Authors: | Mohanram, H. | | Deposit date: | 2019-11-25 | | Release date: | 2020-01-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of Megabalanus rosa Cement Protein 20 revealed by multi-dimensional NMR and molecular dynamics simulations.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 374, 2019
|
|
1MYF
 
 | | SOLUTION STRUCTURE OF CARBONMONOXY MYOGLOBIN DETERMINED FROM NMR DISTANCE AND CHEMICAL SHIFT CONSTRAINTS | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Osapay, K, Theriault, Y, Wright, P.E, Case, D.A. | | Deposit date: | 1994-12-02 | | Release date: | 1995-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of carbonmonoxy myoglobin determined from nuclear magnetic resonance distance and chemical shift constraints.
J.Mol.Biol., 244, 1994
|
|
1T0K
 
 | | Joint X-ray and NMR Refinement of Yeast L30e-mRNA complex | | Descriptor: | 5'-R(*G*GP*AP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*GP*UP*C)-3', 5'-R(*GP*AP*CP*CP*GP*GP*AP*GP*UP*GP*UP*CP*C)-3', 60S ribosomal protein L30, ... | | Authors: | Chao, J.A, Williamson, J.R. | | Deposit date: | 2004-04-09 | | Release date: | 2004-07-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Joint X-Ray and NMR Refinement of the Yeast L30e-mRNA Complex
Structure, 12, 2004
|
|
1JIC
 
 | |
1T8V
 
 | |
5MVB
 
 | | Solution structure of a human G-Quadruplex hybrid-2 form in complex with a Gold-ligand. | | Descriptor: | DNA (26-MER), POTASSIUM ION, bis(m2-Oxo)-bis(2-methyl-2,2'-bipyridine)-di-gold(iii) | | Authors: | Wirmer-Bartoschek, J, Jonker, H.R.A, Bendel, L.E, Gruen, T, Bazzicalupi, C, Messori, L, Gratteri, P, Schwalbe, H. | | Deposit date: | 2017-01-16 | | Release date: | 2017-05-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Ligand/Hybrid-2-G-Quadruplex Complex Reveals Rearrangements that Affect Ligand Binding.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
