2CWG
 
 | |
2CX1
 
 | | Crystal structure of a PUA domain (APE0525) from the Aeropyrum pernix K1 (tartrate complex) | | Descriptor: | L(+)-TARTARIC ACID, hypothetical protein APE0525 | | Authors: | Mizohata, E, Morita, S, Nagano, K, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-27 | | Release date: | 2005-12-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a PUA domain (APE0525) from the Aeropyrum pernix K1 (tartrate complex)
To be Published
|
|
1RVC
 
 | |
2CXA
 
 | | Crystal structure of Leucyl/phenylalanyl-tRNA protein transferase from Escherichia coli | | Descriptor: | Leucyl/phenylalanyl-tRNA-protein transferase | | Authors: | Kato-Murayama, M, Bessho, Y, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-28 | | Release date: | 2005-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of leucyl/phenylalanyl-tRNA-protein transferase from Escherichia coli
Protein Sci., 16, 2007
|
|
1RST
 
 | |
2CV8
 
 | |
2CYA
 
 | | Crystal structure of tyrosyl-tRNA synthetase from Aeropyrum pernix | | Descriptor: | SULFATE ION, Tyrosyl-tRNA synthetase | | Authors: | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Murayama, K, Chen, L, Liu, Z.J, Wang, B.C, Kuroishi, C, Kuramitsu, S, Terada, T, Bessho, Y, Shirouzu, M, Sekine, S.I, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2005
|
|
2D0P
 
 | | Structure of diol dehydratase-reactivating factor in nucleotide free form | | Descriptor: | CALCIUM ION, SULFATE ION, diol dehydratase-reactivating factor large subunit, ... | | Authors: | Shibata, N, Mori, K, Hieda, N, Higuchi, Y, Yamanishi, M, Toraya, T. | | Deposit date: | 2005-08-05 | | Release date: | 2006-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Release of a damaged cofactor from a coenzyme B12-dependent enzyme: X-ray structures of diol dehydratase-reactivating factor
Structure, 13, 2005
|
|
1RO7
 
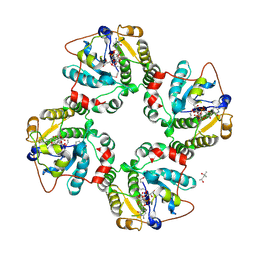 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, CMP-3FNeuAc. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, alpha-2,3/8-sialyltransferase | | Authors: | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2D0A
 
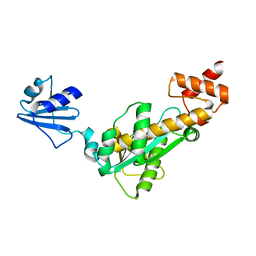 | | Crystal structure of Bst-RNase HIII | | Descriptor: | ribonuclease HIII | | Authors: | Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2005-07-31 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus
J.Mol.Biol., 356, 2006
|
|
2D0O
 
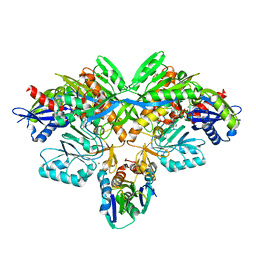 | | Structure of diol dehydratase-reactivating factor complexed with ADP and Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Shibata, N, Mori, K, Hieda, N, Higuchi, Y, Yamanishi, M, Toraya, T. | | Deposit date: | 2005-08-05 | | Release date: | 2006-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Release of a damaged cofactor from a coenzyme B12-dependent enzyme: X-ray structures of diol dehydratase-reactivating factor
Structure, 13, 2005
|
|
1RIS
 
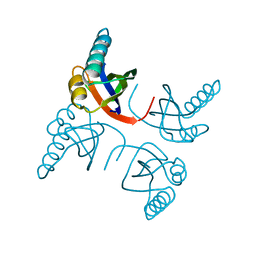 | | CRYSTAL STRUCTURE OF THE RIBOSOMAL PROTEIN S6 FROM THERMUS THERMOPHILUS | | Descriptor: | RIBOSOMAL PROTEIN S6 | | Authors: | Lindahl, M, Svensson, L.A, Liljas, A, Sedelnikova, S.E, Eliseikina, I.A, Fomenkova, N.P, Nevskaya, N, Nikonov, S.V, Garber, M.B, Muranova, T.A, Rykonova, A.I, Amons, R. | | Deposit date: | 1994-05-31 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the ribosomal protein S6 from Thermus thermophilus.
EMBO J., 13, 1994
|
|
1ZBF
 
 | | Crystal structure of B. halodurans RNase H catalytic domain mutant D132N | | Descriptor: | SULFATE ION, ribonuclease H-related protein | | Authors: | Nowotny, M, Gaidamakov, S.A, Crouch, R.J, Yang, W. | | Deposit date: | 2005-04-08 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis.
Cell(Cambridge,Mass.), 121, 2005
|
|
2CVK
 
 | |
2CYC
 
 | | Crystal structure of Tyrosyl-tRNA Synthetase complexed with L-tyrosine from Pyrococcus horikoshii | | Descriptor: | TYROSINE, tyrosyl-tRNA synthetase | | Authors: | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Sakamoto, K, Terada, T, Shirouzu, M, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2006
|
|
1Z1F
 
 | | Crystal structure of stilbene synthase from Arachis hypogaea (resveratrol-bound form) | | Descriptor: | CITRIC ACID, RESVERATROL, stilbene synthase | | Authors: | Shomura, Y, Torayama, I, Suh, D.Y, Xiang, T, Kita, A, Sankawa, U, Miki, K. | | Deposit date: | 2005-03-03 | | Release date: | 2005-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of stilbene synthase from Arachis hypogaea
Proteins, 60, 2005
|
|
2CIU
 
 | |
1RVK
 
 | | Crystal structure of enolase AGR_L_2751 from Agrobacterium Tumefaciens | | Descriptor: | MAGNESIUM ION, isomerase/lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Thirumuruhan, R, Zencheck, W, Millikin, C, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-12-14 | | Release date: | 2003-12-23 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of enzymatic activites in the Enolase superfamily: 1.7 A crystal structure of the hypothetical protein MR.GI-17937161 from Agrobacterium tumefaciens
To be Published
|
|
1ZHX
 
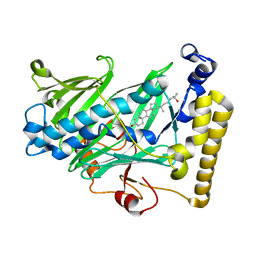 | | Structure of yeast oxysterol binding protein Osh4 in complex with 25-hydroxycholesterol | | Descriptor: | 25-HYDROXYCHOLESTEROL, KES1 protein | | Authors: | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
1ZI9
 
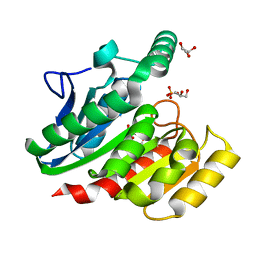 | | Crystal Structure Analysis of the dienelactone hydrolase (E36D, C123S) mutant- 1.5 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1RNM
 
 | |
1ROA
 
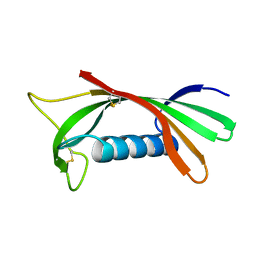 | | Structure of human cystatin D | | Descriptor: | Cystatin D | | Authors: | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | Deposit date: | 2003-12-01 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
1Z33
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase | | Descriptor: | purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1ZJ4
 
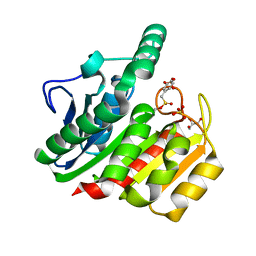 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (E36D, C123S) bound with the PMS moiety of the protease inhibitor, Phenylmethylsulfonyl fluoride (PMSF)- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Z39
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2'-deoxyinosine | | Descriptor: | 2'-DEOXYINOSINE, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
