1QJ4
 
 | |
3YAS
 
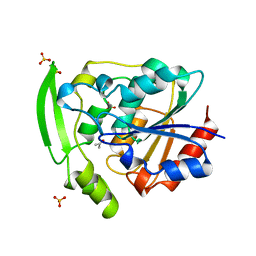 | | HYDROXYNITRILE LYASE COMPLEXED WITH ACETONE | | Descriptor: | ACETONE, PROTEIN (HYDROXYNITRILE LYASE), SULFATE ION | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
1BI9
 
 | | RETINAL DEHYDROGENASE TYPE TWO WITH NAD BOUND | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, RETINAL DEHYDROGENASE TYPE II | | Authors: | Newcomer, M.E, Lamb, A.L. | | Deposit date: | 1998-06-23 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of retinal dehydrogenase type II at 2.7 A resolution: implications for retinal specificity.
Biochemistry, 38, 1999
|
|
3DMV
 
 | |
3DN2
 
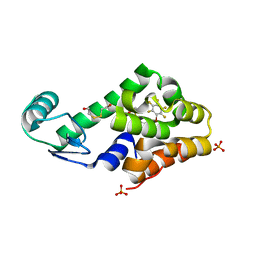 | | Bromopentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant | | Descriptor: | 1-bromo-2,3,4,5,6-pentafluorobenzene, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
3DN8
 
 | | Iodopentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant (seleno version) | | Descriptor: | 1,2,3,4,5-pentafluoro-6-iodobenzene, 2-HYDROXYETHYL DISULFIDE, BETA-MERCAPTOETHANOL, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
3DN6
 
 | | 1,3,5-trifluoro-2,4,6-trichlorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant | | Descriptor: | 1,3,5-trichloro-2,4,6-trifluorobenzene, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
1MEG
 
 | |
2OVW
 
 | | ENDOGLUCANASE I COMPLEXED WITH CELLOBIOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Sulzenbacher, G, Davies, G.J, Schulein, M. | | Deposit date: | 1997-04-04 | | Release date: | 1998-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the endoglucanase I from Fusarium oxysporum: native, cellobiose, and 3,4-epoxybutyl beta-D-cellobioside-inhibited forms, at 2.3 A resolution.
Biochemistry, 36, 1997
|
|
3DMX
 
 | |
3DN3
 
 | | Iodopentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant | | Descriptor: | 1,2,3,4,5-pentafluoro-6-iodobenzene, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
4B2V
 
 | | S64, a spider venom toxin peptide from Sicarius dolichocephalus | | Descriptor: | S64 | | Authors: | Loening, N.M, Wilson, Z.N, Zobel-Thropp, P.A, Binford, G.J. | | Deposit date: | 2012-07-18 | | Release date: | 2013-01-16 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Two Homologous Venom Peptides from Sicarius Dolichocephalus
Plos One, 8, 2013
|
|
3DKE
 
 | | Polar and non-polar cavities in phage T4 lysozyme | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AZIDE ION, ... | | Authors: | Liu, L.J, Matthews, B.W. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Use of experimental crystallographic phases to examine the hydration of polar and nonpolar cavities in T4 lysozyme
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1ABR
 
 | | CRYSTAL STRUCTURE OF ABRIN-A | | Descriptor: | ABRIN-A, beta-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-L-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, beta-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Tahirov, T.H, Lu, T.-H, Liaw, Y.-C, Chu, S.-C, Lin, J.-Y. | | Deposit date: | 1994-11-11 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of abrin-a at 2.14 A.
J.Mol.Biol., 250, 1995
|
|
1NAR
 
 | |
1NCE
 
 | | Crystal structure of a ternary complex of E. coli thymidylate synthase D169C with dUMP and the antifolate CB3717 | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Birdsall, D.L, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2002-12-05 | | Release date: | 2002-12-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The only active mutant of thymidylate synthase D169, a residue far from the
site of methyl transfer, demonstrates the exquisite nature of enzyme
specificity.
Protein Eng., 16, 2003
|
|
1RIE
 
 | | STRUCTURE OF A WATER SOLUBLE FRAGMENT OF THE RIESKE IRON-SULFUR PROTEIN OF THE BOVINE HEART MITOCHONDRIAL CYTOCHROME BC1-COMPLEX | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, RIESKE IRON-SULFUR PROTEIN | | Authors: | Iwata, S, Saynovits, M, Link, T.A, Michel, H. | | Deposit date: | 1996-02-23 | | Release date: | 1996-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a water soluble fragment of the 'Rieske' iron-sulfur protein of the bovine heart mitochondrial cytochrome bc1 complex determined by MAD phasing at 1.5 A resolution.
Structure, 4, 1996
|
|
1WGK
 
 | | Solution Structure of Mouse Hypothetical Protein 2900073H19RIK | | Descriptor: | RIKEN cDNA 2900073H19 protein | | Authors: | Zhao, C, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mouse Hypothetical Protein 2900073H19RIK
To be Published
|
|
1WHJ
 
 | | Solution structure of the 1st CAP-Gly domain in mouse 1700024K14Rik hypothetical protein | | Descriptor: | RIKEN cDNA 1700024K14 | | Authors: | Saito, K, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 1st CAP-Gly domain in mouse 1700024K14Rik hypothetical protein
To be Published
|
|
1WHN
 
 | | Solution structure of the dsRBD from hypothetical protein BAB26260 | | Descriptor: | hypothetical protein RIKEN cDNA 2310016K04 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dsRBD from hypothetical protein BAB26260
To be Published
|
|
1WEX
 
 | | Solution structure of RRM domain in protein BAB28521 | | Descriptor: | HYPOTHETICAL PROTEIN (RIKEN CDNA 2810036L13) | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in protein BAB28521
To be Published
|
|
1WHW
 
 | | Solution structure of the N-terminal RNA binding domain from hypothetical protein BAB23448 | | Descriptor: | hypothetical protein RIKEN CDNA 1200009A02 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal RNA binding domain from hypothetical protein BAB23448
To be Published
|
|
1WHX
 
 | | Solution structure of the second RNA binding domain from hypothetical protein BAB23448 | | Descriptor: | hypothetical protein RIKEN CDNA 1200009A02 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RNA binding domain from hypothetical protein BAB23448
To be Published
|
|
1WIF
 
 | | The solution structure of RSGI RUH-020, a PDZ domain of hypothetical protein from mouse | | Descriptor: | RIKEN cDNA 4930408O21 | | Authors: | Ohashi, W, Yamazaki, T, Hirota, H, Tomizawa, T, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of RSGI RUH-020, a PDZ domain of hypothetical protein from mouse
To be Published
|
|
1WHY
 
 | | Solution structure of the RNA recognition motif from hypothetical RNA binding protein BC052180 | | Descriptor: | hypothetical protein RIKEN cDNA 1810017N16 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA recognition motif from hypothetical RNA binding protein BC052180
To be Published
|
|
