6OD1
 
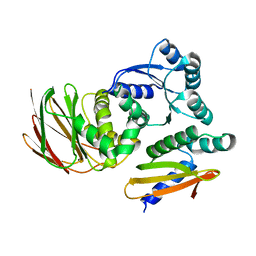 | | IraD-bound to RssB D58P variant | | Descriptor: | Anti-adapter protein IraD, Regulator of RpoS | | Authors: | Deaconescu, A.M, Dorich, V. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of a response regulator of sigmaSstability by a ClpXP antiadaptor.
Genes Dev., 33, 2019
|
|
6QYQ
 
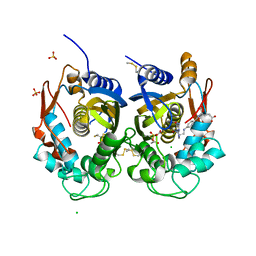 | | Crystal structure of human thymidylate synthase (hTS) variant R175C | | Descriptor: | CHLORIDE ION, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SULFATE ION, ... | | Authors: | Pozzi, C, Mangani, M. | | Deposit date: | 2019-03-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and Functional Characterization of the Human Thymidylate Synthase (hTS) Interface Variant R175C, New Perspectives for the Development of hTS Inhibitors.
Molecules, 24, 2019
|
|
1Y6U
 
 | |
6OLA
 
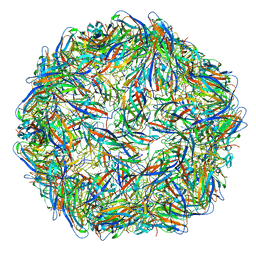 | | Structure of the PCV2d virus-like particle | | Descriptor: | Capsid protein, DNA (5'-D(P*CP*CP*GP*G)-3') | | Authors: | Khayat, R, Wen, K, Alimova, A, Galarza, J, Gottlieb, P. | | Deposit date: | 2019-04-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural characterization of the PCV2d virus-like particle at 3.3 angstrom resolution reveals differences to PCV2a and PCV2b capsids, a tetranucleotide, and an N-terminus near the icosahedral 3-fold axes.
Virology, 537, 2019
|
|
1JNX
 
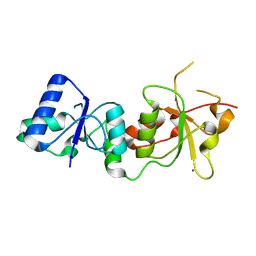 | |
6QXH
 
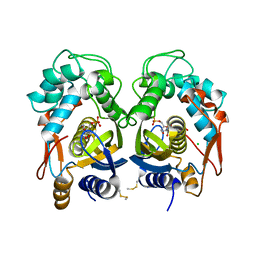 | |
6QXG
 
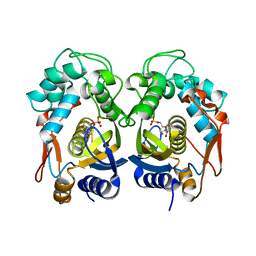 | |
1KEY
 
 | | Crystal Structure of Mouse Testis/Brain RNA-binding Protein (TB-RBP) | | Descriptor: | translin | | Authors: | Pascal, J.M, Hart, P.J, Hecht, N.B, Robertus, J.D. | | Deposit date: | 2001-11-19 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of TB-RBP, a Novel RNA-binding and Regulating Protein
J.Mol.Biol., 319, 2002
|
|
1S9H
 
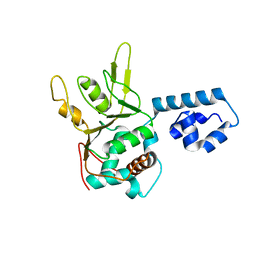 | | Crystal Structure of Adeno-associated virus Type 2 Rep40 | | Descriptor: | Rep 40 protein | | Authors: | James, J.A, Escalante, C.R, Yoon-Robarts, M, Edwards, T.A, Linden, R.M, Aggarwal, A.K. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the SF3 Helicase from Adeno-Associated Virus Type 2
Structure, 11, 2003
|
|
6R2E
 
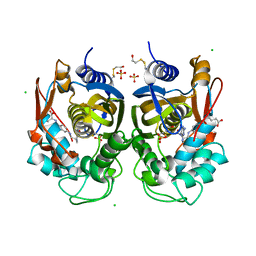 | | Crystal structure of the human thymidylate synthase (hTS) interface variant Q62R | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Pozzi, C, Mangani, M. | | Deposit date: | 2019-03-16 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Evidence of Destabilization of the Human Thymidylate Synthase (hTS) Dimeric Structure Induced by the Interface Mutation Q62R.
Biomolecules, 9, 2019
|
|
1OFC
 
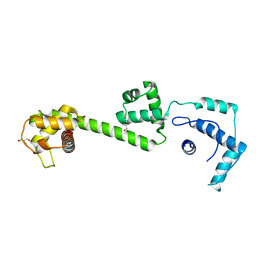 | | nucleosome recognition module of ISWI ATPase | | Descriptor: | 4-deoxy-alpha-D-glucopyranose, GLYCEROL, ISWI PROTEIN, ... | | Authors: | Grune, T, Muller, C.W. | | Deposit date: | 2003-04-10 | | Release date: | 2003-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Functional Analysis of a Nucleosome Recognition Module of the Remodeling Factor Iswi
Mol.Cell, 12, 2003
|
|
3JSC
 
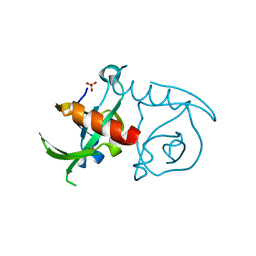 | | CcdBVfi-FormI-pH7.0 | | Descriptor: | CcdB, SULFATE ION | | Authors: | De Jonge, N, Buts, L, Loris, R. | | Deposit date: | 2009-09-10 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and thermodynamic characterization of vibrio fischeri CCDB
J.Biol.Chem., 285, 2010
|
|
1JRN
 
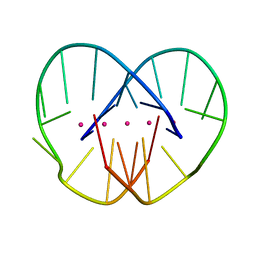 | |
1SVL
 
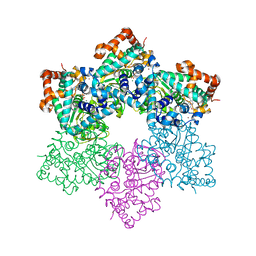 | | Co-crystal structure of SV40 large T antigen helicase domain and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
1L6B
 
 | |
1NCO
 
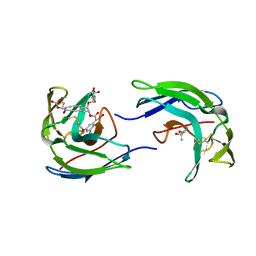 | | STRUCTURE OF THE ANTITUMOR PROTEIN-CHROMOPHORE COMPLEX NEOCARZINOSTATIN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, HOLO-NEOCARZINOSTATIN, NEOCARZINOSTATIN-CHROMOPHORE | | Authors: | Kim, K.-H, Kwon, B.-M, Myers, A.G, Rees, D.C. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of neocarzinostatin, an antitumor protein-chromophore complex.
Science, 262, 1993
|
|
1J74
 
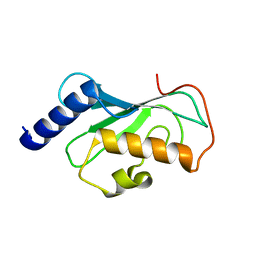 | | Crystal Structure of Mms2 | | Descriptor: | MMS2 | | Authors: | Moraes, T.F, Edwards, R.A, McKenna, S, Pastushok, L, Xiao, W, Glover, J.N.M, Ellison, M.J. | | Deposit date: | 2001-05-15 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13.
Nat.Struct.Biol., 8, 2001
|
|
4CS8
 
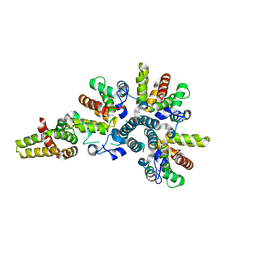 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer, form 2 | | Descriptor: | M2-1, ZINC ION | | Authors: | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
4CS7
 
 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer, form 1 | | Descriptor: | M2-1, ZINC ION | | Authors: | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
4CS9
 
 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer bound to adenosine monophosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, M2-1, ZINC ION | | Authors: | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
7QOF
 
 | | Icosahedral capsid of the phicrAss001 virion | | Descriptor: | Auxiliary capsid protein gp36, Head fiber dimer protein gp29, Head fiber trimer protein gp21, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOH
 
 | | Unique vertex of the phicrAss001 virion with C5 symmetry imposed | | Descriptor: | Auxiliary capsid protein gp36, Head fiber trimer protein gp21, MAGNESIUM ION, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOG
 
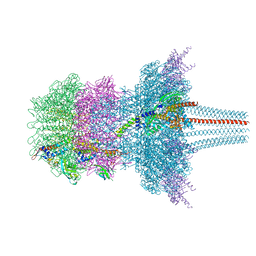 | | Portal protein assembly of the phicrAss001 virion with C12 symmetry imposed | | Descriptor: | Cargo protein 1 gp45, Portal protein gp20, Ring protein 1 gp43, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOK
 
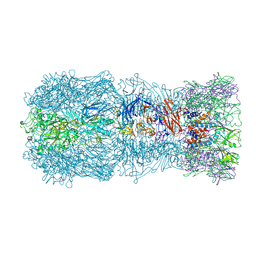 | | Tail muzzle assembly of the phicrAss001 virion with C6 symmetry imposed | | Descriptor: | MAGNESIUM ION, Muzzle bound helix, Muzzle protein gp44, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOJ
 
 | | Tail barrel assembly of the phicrAss001 virion with C12 symmetry imposed | | Descriptor: | Cargo protein 1 gp45, MAGNESIUM ION, Portal protein gp20, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
