2AO6
 
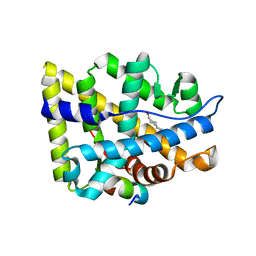 | | Crystal structure of the human androgen receptor ligand binding domain bound with TIF2(iii) 740-753 peptide and R1881 | | Descriptor: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, 14-mer fragment of Nuclear receptor coactivator 2, androgen receptor | | Authors: | He, B, Gampe Jr, R.T, Kole, A.J, Hnat, A.T, Stanley, T.B, An, G, Stewart, E.L, Kalman, R.I, Minges, J.T, Wilson, E.M. | | Deposit date: | 2005-08-12 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance
Mol.Cell, 16, 2004
|
|
6G99
 
 | | Solution structure of FUS-ZnF bound to UGGUG | | Descriptor: | RNA (5'-R(*UP*GP*GP*UP*G)-3'), RNA-binding protein FUS, ZINC ION | | Authors: | Loughlin, F.E, Allain, F.H.-T. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity.
Mol. Cell, 73, 2019
|
|
6FW7
 
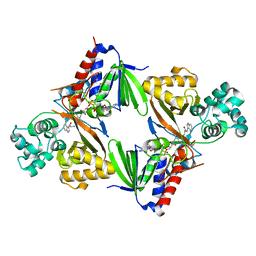 | | Crystal structure of L-tryptophan oxidase VioA from Chromobacterium violaceum in complex with 4-Fluoro-L-Tryptophan | | Descriptor: | 4-FLUOROTRYPTOPHANE, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA, ... | | Authors: | Lai, H.E, Morgan, M, Moore, S, Freemont, P. | | Deposit date: | 2018-03-05 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A GenoChemetic strategy for derivatization of the violacein natural product scaffold
Biorxiv, 2019
|
|
6G9R
 
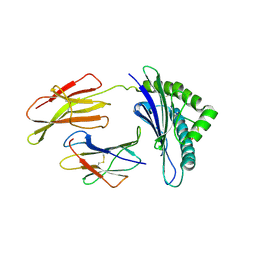 | |
8DMI
 
 | | Lymphocytic choriomeningitis virus glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein G1, ... | | Authors: | Moon-Walker, A, Hastie, K.M, Zyla, D.S, Saphire, E.O. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for antibody-mediated neutralization of lymphocytic choriomeningitis virus.
Cell Chem Biol, 30, 2023
|
|
4TU4
 
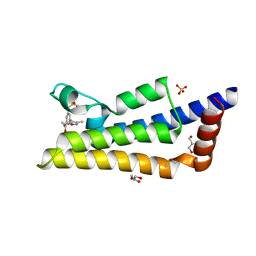 | | Crystal structure of ATAD2A bromodomain complexed with 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(phenylsulfonyl)amino]benzoicacid | | Descriptor: | 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(phenylsulfonyl)amino]benzoic acid, ATPase family AAA domain-containing protein 2, CHLORIDE ION, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
6FXT
 
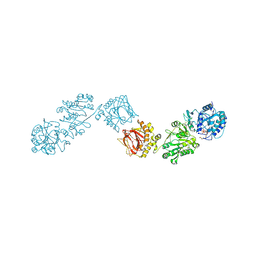 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Glc | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scietti, L, Chiapparino, A, De Giorgi, F, Fumagalli, M, Khoriauli, L, Nergadze, S, Basu, S, Olieric, V, Banushi, B, Giulotto, E, Gissen, P, Forneris, F. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun, 9, 2018
|
|
3ZE3
 
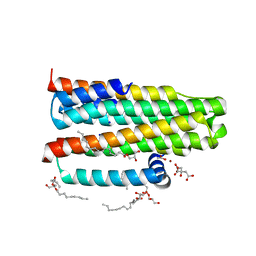 | | Crystal structure of the integral membrane diacylglycerol kinase - delta7 | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ACETATE ION, ... | | Authors: | Li, D, Pye, V.E, Lyons, J.A, Vogeley, L, Aragao, D, Caffrey, M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the Integral Membrane Diacylglycerol Kinase.
Nature, 497, 2013
|
|
5TLV
 
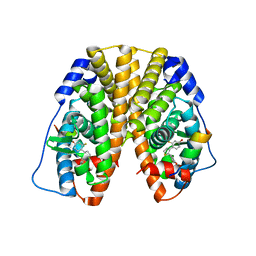 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 4,4'-(thiophene-2,3-diyl)bis(3-fluorophenol) | | Descriptor: | 4,4'-(thiene-2,3-diyl)bis(3-fluorophenol), Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.323 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TLY
 
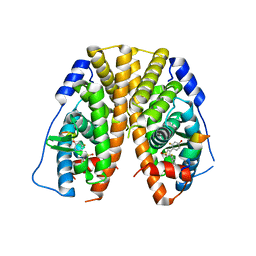 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 3,4-bis(2-fluoro-4-hydroxyphenyl)thiophene 1,1-dioxide | | Descriptor: | 3,4-bis(2-fluoro-4-hydroxyphenyl)-1H-1lambda~6~-thiophene-1,1-dione, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM4
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC Analog, 5-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)pentanoic acid | | Descriptor: | 5-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}pentanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM7
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 7-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)heptanoic acid | | Descriptor: | 7-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}heptanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMW
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS derivative, 4-acetamidophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | Descriptor: | 4-(acetylamino)phenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.286 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TN3
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the estradiol derivative, (8S,9S,13S,14S)-17-((4-isopropylphenyl)amino)-13-methyl-7,8,9,11,12,13,14,15,16,17-decahydro-6H-cyclopenta[a]phenanthren-3-ol | | Descriptor: | (9beta,13alpha,17beta)-17-{[4-(propan-2-yl)phenyl]amino}estra-1(10),2,4-trien-3-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMU
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 4,4'-(cycloheptylidenemethylene)diphenol | | Descriptor: | 4,4'-(cycloheptylidenemethylene)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.429 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TN4
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the ACD-ring estrogen, (S)-5-(4-hydroxy-3,5-dimethylphenyl)-2,3-dihydro-1H-inden-1-ol | | Descriptor: | (1S)-5-(4-hydroxy-3,5-dimethylphenyl)-2,3-dihydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TO7
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
6QFY
 
 | | CRYSTAL STRUCTURE OF PORCINE HEMAGGLUTINATING ENCEPHALOMYELITIS VIRUS SPIKE PROTEIN LECTIN DOMAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Huizinga, E.G, Bakkers, M, Lang, Y. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6VWY
 
 | |
4A22
 
 | | Structure of Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase bound to N-(4-hydroxybutyl)- glycolohydroxamic acid bis- phosphate | | Descriptor: | 4-{hydroxy[(phosphonooxy)acetyl]amino}butyl dihydrogen phosphate, FRUCTOSE-BISPHOSPHATE ALDOLASE, SODIUM ION, ... | | Authors: | Coincon, M, De la Paz Santangelo, M, Gest, P.M, Guerin, M.E, Pham, H, Ryan, G, Puckett, S.E, Spencer, J.S, Gonzalez-Juarrero, M, Daher, R, Lenaerts, A.J, Schnappinger, D, Therisod, M, Ehrt, S, Jackson, M, Sygusch, J. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Glycolytic and non-glycolytic functions of Mycobacterium tuberculosis fructose-1,6-bisphosphate aldolase, an essential enzyme produced by replicating and non-replicating bacilli.
J. Biol. Chem., 286, 2011
|
|
2JLD
 
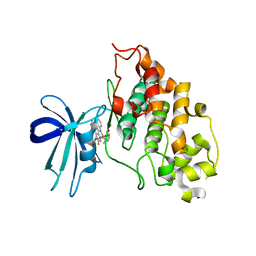 | | Extremely Tight Binding of Ruthenium Complex to Glycogen Synthase Kinase 3 | | Descriptor: | GLYCOGEN SYNTHASE KINASE-3 BETA, PEPTIDE (ALA-GLY-GLY-ALA-ALA-ALA-ALA-ALA), RUTHENIUM PYRIDOCARBAZOLE | | Authors: | Atilla-Gokcumen, G.E, Pagano, N, Streu, C, Maksimoska, J, Filippakopoulos, P, Knapp, S, Meggers, E. | | Deposit date: | 2008-09-08 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Extremely Tight Binding of a Ruthenium Complex to Glycogen Synthase Kinase 3.
Chembiochem, 9, 2008
|
|
4K1P
 
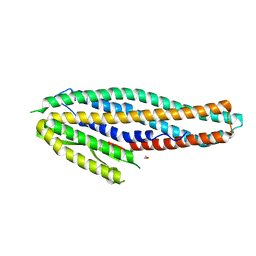 | | Structure of the NheA component of the Nhe toxin from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, NheA, SULFATE ION | | Authors: | Ganash, M, Phung, D, Artymiuk, P.J. | | Deposit date: | 2013-04-05 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the NheA Component of the Nhe Toxin from Bacillus cereus: Implications for Function.
Plos One, 8, 2013
|
|
5TM8
 
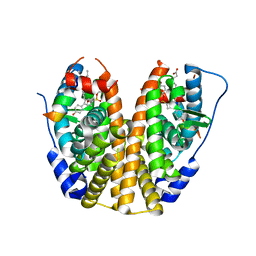 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 7-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)heptanoic acid | | Descriptor: | 7-{4-[(1S,4S,6R)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}heptanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
8E5F
 
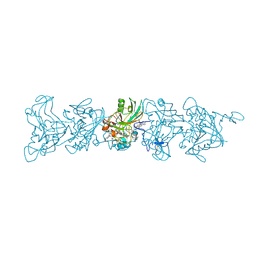 | | Cryo-EM of P. calidifontis cytochrome filament | | Descriptor: | HEME C, c-type cytochrome | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-10 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Extracellular cytochrome nanowires appear to be ubiquitous in prokaryotes.
Cell, 186, 2023
|
|
2X42
 
 | |
